|
Maturase K
Maturase K (matK) is a plant plastidial gene. The protein it encodes is an organelle intron maturase, a protein that splices Group II introns. It is essential for ''in vivo'' splicing of Group II introns. Amongst other maturases, this protein retains only a well conserved domain X and remnants of a reverse transcriptase domain. Universal matK primers can be used for DNA barcoding of angiosperms. See also * LtrA, an open reading frame In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames ... found in the ''Lactococcus lactis'' group II introns LtrB. It is an intron-encoded protein, with three subdomains, one of which is a reverse-transcriptase/maturase. References Plant genes Plastids {{gene-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arabidopsis Thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed. A winter annual with a relatively short lifecycle, ''A. thaliana'' is a popular model organism in plant biology and genetics. For a complex multicellular eukaryote, ''A. thaliana'' has a relatively small genome of around 135 Base pair#Length measurements, megabase pairs. It was the first plant to have its genome sequenced, and is an important tool for understanding the molecular biology of many plant traits, including flower development and phototropism, light sensing. Description ''Arabidopsis thaliana'' is an annual plant, annual (rarely biennial plant, biennial) plant, usually growing to 20–25 cm tall. The leaf, leaves form a rosette at the base of the plant, with a few leaves also on the flowering Plant ste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
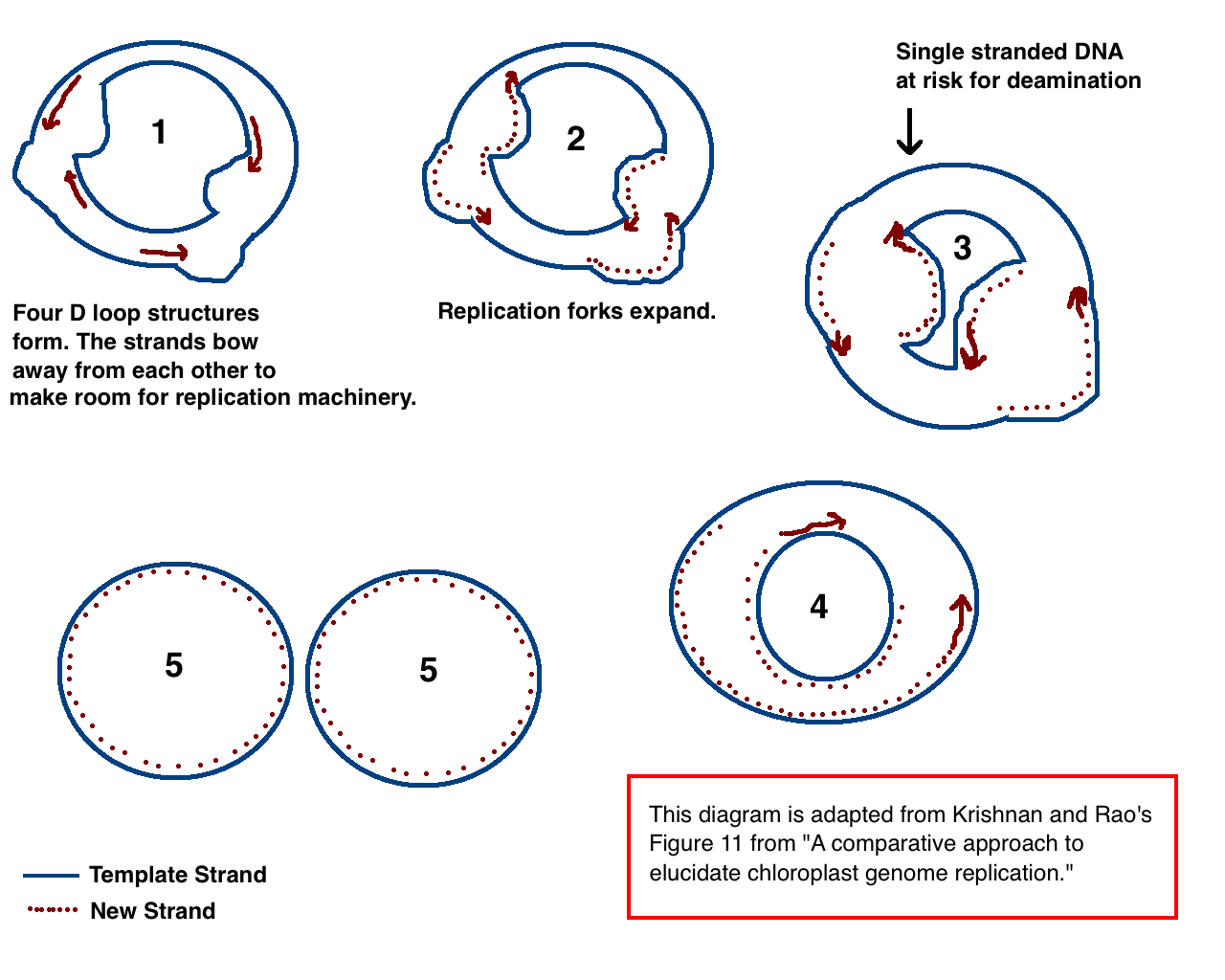
Chloroplast DNA
Chloroplast DNA (cpDNA), also known as plastid DNA (ptDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, ''Nicotiana tabacum'' (tobacco) by Sugiura and colleagues and '' Marchantia polymorpha'' (liverwort) by Ozeki et al. Since then, tens of thousands of chloroplast genomes from various species have been sequenced. Molecular structure Chloroplast DNAs are circular, and are typically 120,000–170,000 base pairs long. They can have a contour length of around 30–60 mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear genes, nuclear-encoded genes, splicing occurs in the cell nucleus, nucleus either during or immediately after Transcription (biology), transcription. For those eukaryotic transcription, eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translation (biology), translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Group II Intron
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity (e.g., self- splicing) can occur under high-salt conditions ''in vitro''. However, assistance from proteins is required for ''in vivo'' splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing (see spliceosome) may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain X
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three Domain (biology), domains of life. Ribozyme activity (e.g., self-RNA splicing, splicing) can occur under high-salt conditions ''in vitro''. However, assistance from proteins is required for ''in vivo'' splicing. In contrast to Group I catalytic intron, group I introns, intron excision occurs in the absence of Guanosine triphosphate, GTP and involves the formation of a Lariat (other), lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing (see spliceosome) may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended Small nuclear RNA, snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. The process does not violate the flows of genetic information as described by the classical central dogma, but rather expands it to include transfers of information from RNA to DNA. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primer (molecular Biology)
A primer is a short, single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer is a type of oligo, short for oligonucleotide. DNA polymerases (responsible for DNA replication) are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replaced with DNA nucleotides. This forms a gap region known as a nick that is filled in using a ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called " sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the cytochrome ''c'' oxidase I (COI or COX1) gene, found in mitochondrial DNA. Other genes suitable for DNA barcoding a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Angiosperm
Flowering plants are plants that bear flowers and fruits, and form the clade Angiospermae (). The term angiosperm is derived from the Greek words (; 'container, vessel') and (; 'seed'), meaning that the seeds are enclosed within a fruit. The group was formerly called Magnoliophyta. Angiosperms are by far the most diverse group of land plants with 64 orders, 416 families, approximately 13,000 known genera and 300,000 known species. They include all forbs (flowering plants without a woody stem), grasses and grass-like plants, a vast majority of broad-leaved trees, shrubs and vines, and most aquatic plants. Angiosperms are distinguished from the other major seed plant clade, the gymnosperms, by having flowers, xylem consisting of vessel elements instead of tracheids, endosperm within their seeds, and fruits that completely envelop the seeds. The ancestors of flowering plants diverged from the common ancestor of all living gymnosperms before the end of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LtrA
''LtrA'' is an open reading frame found in the '' Lactococcus lactis'' group II introns LtrB. It is an intron-encoded protein, which consists of three subdomains: a reverse-transcriptase/ maturase, DNA endonuclease, and DNA/RNA binding domain In molecular biology, binding domain is a protein domain which binds to a specific atom or molecule, such as calcium or DNA. A protein domain is a part of a protein sequence and a tertiary structure that can change or evolve, function, and live .... LtrA helps to capture and stabilize the catalytically active conformation of the LtrB group II intron RNA. It also functions in group II intron retrohoming. References Prokaryote genes {{molecular-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |