|
Mannosyl-glycoprotein Endo-b-N-acetylglucosaminidase
The enzyme mannosyl-glycoprotein endo-β-''N''-acetylglucosaminidase (endoglycosidase H) () has systematic name ''glycopeptide-D-mannosyl-N4-(N-acetyl-D-glucosaminyl)2-asparagine 1,4-N-acetyl-β-glucosaminohydrolase''. It is a highly specific endoglycosidase which cleaves asparagine-linked mannose rich oligosaccharides, but not highly processed complex oligosaccharides from glycoproteins. It is used for research purposes to deglycosylate glycoproteins and to monitor intracellular protein trafficking through the secretory pathway. Structure and activity Endoglycosidase H is isolated from ''Streptomyces plicatus'' or ''Streptomyces griseus''. Its molecular mass is 29 kDa. The primary structure was described by Robbins et al. in 1984. Endoglycosidase H cleaves the bond in the diacetylchitobiose core of the oligosaccharide between two ''N''-acetylglucosamine (GlcNAc) subunits directly proximal to the asparagine residue, generating a truncated sugar molecule with one ''N''-acetylglu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
Enzymes are listed here by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system: :Oxidoreductases (EC 1) ( Oxidoreductase) * Dehydrogenase * Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase ** Diacetyl reductase ** Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase ** glycerol-3-phoshitiendopene dehydrogenase (NAD+) ** D-xylulose reductase ** L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * EC 1.1.4 (with a disulfide as accep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Posttranslational Modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can expand the chemical set of the 22 amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosylation, which can promote protein folding and improve stability a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoglycosidase
An Endoglycosidase is an enzyme that releases oligosaccharides from glycoproteins or glycolipids. It may also cleave polysaccharide chains between residues that are not the terminal residue, although releasing oligosaccharides from conjugated protein and lipid molecules is more common. It breaks the glycosidic bonds between two sugar monomer in the polymer. It is different from exoglycosidase that it does not do so at the terminal residue. Hence, it is used to release long carbohydrates from conjugated molecules. If an exoglycosidase were used, every monomer in the polymer would have to be removed, one by one from the chain, taking a long time. An endoglycosidase cleaves, giving a polymeric product. PROTEIN-x1-x2-x3-x4-x5-x6-x7-x8-x9-x10-x11-...-xn Mechanism Overview The mechanism is an enzymatic hydrolysis that requires two critical molecules; a proton donor (most likely an acid) and a nucleophile(most likely a base). The Endoglycosidases mechanism has two forms; an acid cataly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mannose
Mannose is a sugar with the formula , which sometimes is abbreviated Man. It is one of the monomers of the aldohexose series of carbohydrates. It is a C-2 epimer of glucose. Mannose is important in human metabolism, especially in the glycosylation of certain proteins. Several congenital disorders of glycosylation are associated with mutations in enzymes involved in mannose metabolism. Mannose is not an essential nutrient; it can be produced in the human body from glucose, or converted into glucose. Mannose provides 2–5 kcal/g. It is partially excreted in the urine. Etymology The root of both "mannose" and " mannitol" is manna, which the Bible describes as the food supplied to the Israelites during their journey in the region of Sinai. Several trees and shrubs can produce a substance called manna, such as the "manna tree" (''Fraxinus ornus'') from whose secretions mannitol was originally isolated. Structure Mannose commonly exists as two different-sized rings, the py ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-mannosidase II
Mannosyl-oligosaccharide 1,3-1,6-α-mannosidase, also known as Golgi α-mannosidase II, is an enzyme with systematic name ''(1→3)-(1→6)-mannosyl-oligosaccharide α-D-mannohydrolase''. It catalyses the hydrolysis of the terminal (1→3)- and (1→6)-linked α-D-mannose residues in the mannosyl-oligosaccharide Man5(GlcNAc)3. This enzyme is involved in the synthesis of glycoproteins. It is a key enzyme of ''N''-linked glycan processing and is inhibited by small molecule swainsonine Swainsonine is an indolizidine alkaloid. It is a potent inhibitor of Golgi alpha-mannosidase II, an immunomodulator, and a potential chemotherapy drug. As a toxin in locoweed (likely its primary toxin) it also is a significant cause of econo .... References External links * {{Portal bar, Biology, border=no EC 3.2.1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligosaccharyl Transferase
Oligosaccharyltransferase or OST () is a membrane protein complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc= ''N''-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X- Ser or Asn-X- Thr where X is any amino acid except proline. This sequence is called a glycosylation ''sequon.'' The reaction catalyzed by OST is the central step in the ''N''-linked glycosylation pathway. Location OST is a component of the translocon in the endoplasmic reticulum (ER) membrane. A lipid-linked core-oligosaccharide is assembled at the membrane of the endoplasmic reticulum and transferred to selected asparagine residues of nascent polypeptide chains by the oligosaccharyl transferase complex. The active site of OST is located about 4 nm from the lumenal face of the ER membrane. It usually acts during translation as the nas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is a part of a transportation system of the eukaryote, eukaryotic cell, and has many other important functions such as protein folding. The word endoplasmic means "within the cytoplasm", and reticulum is Latin for "little net". It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. There are two types of ER that share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of Cell (biology), cells contain different ratios of the two types of ER dependin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomes
Ribosomes () are macromolecular machines, found within all cells, that perform biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins (). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA (mRNA) chain. Ribosomes bind to the messenger RNA molecules and use the RNA's sequence of nucleotides to determine the sequence of amino acids needed to generate a protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anticodo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (genetics)
In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in the addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The polypeptide can also start folding during protein synthesis. The ribosome facilitates decoding by inducing the binding of complementary transfe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
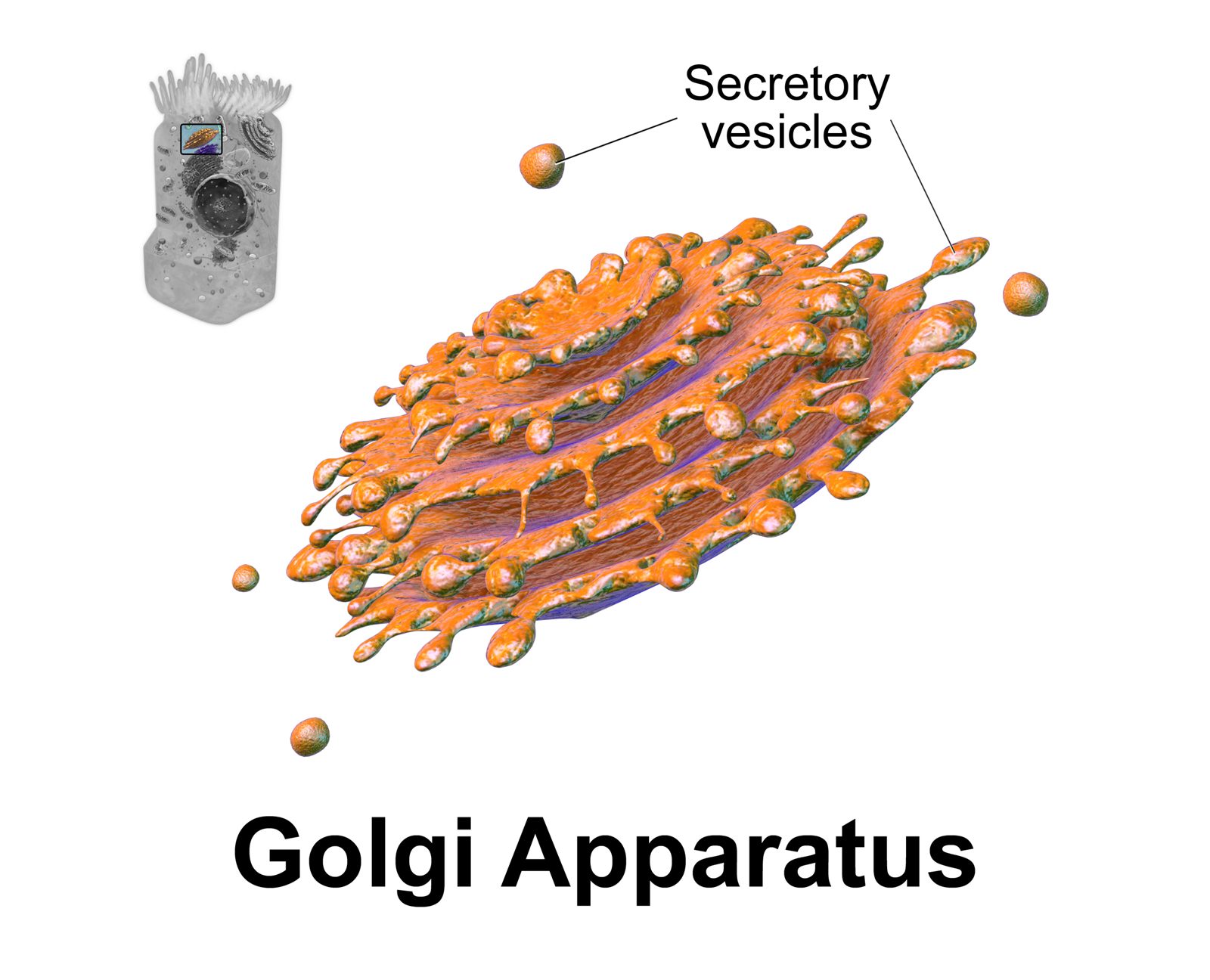
Golgi Apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic Cell (biology), cells. Part of the endomembrane system in the cytoplasm, it protein targeting, packages proteins into membrane-bound Vesicle (biology and chemistry), vesicles inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and Endocytosis, endocytic pathways. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. The Golgi apparatus was identified in 1898 by the Italian biologist and pathologist Camillo Golgi. The organelle was later named after him in the 1910s. Discovery Because of its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoglycosidase
An Endoglycosidase is an enzyme that releases oligosaccharides from glycoproteins or glycolipids. It may also cleave polysaccharide chains between residues that are not the terminal residue, although releasing oligosaccharides from conjugated protein and lipid molecules is more common. It breaks the glycosidic bonds between two sugar monomer in the polymer. It is different from exoglycosidase that it does not do so at the terminal residue. Hence, it is used to release long carbohydrates from conjugated molecules. If an exoglycosidase were used, every monomer in the polymer would have to be removed, one by one from the chain, taking a long time. An endoglycosidase cleaves, giving a polymeric product. PROTEIN-x1-x2-x3-x4-x5-x6-x7-x8-x9-x10-x11-...-xn Mechanism Overview The mechanism is an enzymatic hydrolysis that requires two critical molecules; a proton donor (most likely an acid) and a nucleophile(most likely a base). The Endoglycosidases mechanism has two forms; an acid cataly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |