|
LoxP
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community. An important application of the Cre-lox system is excision of selectable markers in gene replacement. Commonly used gene replacement strategies introduce selectable markers into the genome to facilitate selection of genetic mutations that may cause growth retardation. Ho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cre Recombinase
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38 kDa) is a member of the integrase family of site specific recombinase and it is known to catalyse the site specific recombination event between two DNA recognition sites ( LoxP sites). This 34 base pair (bp) loxP recognition site consists of two 13 bp palindromic sequences which flank an 8bp spacer region. The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. DNA sequences found between two loxP sites are said to be " floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites oriented in the same direction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
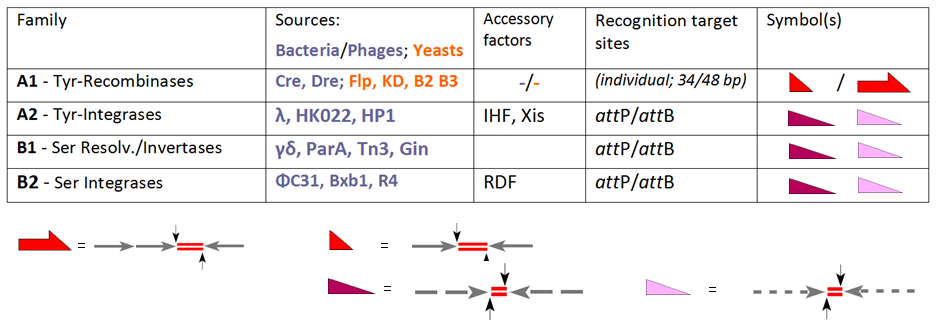
Site-specific Recombinase Technology
Site-specific recombinase technologies are genome engineering tools that depend on Recombinase, recombinase enzymes to replace targeted sections of DNA. History In the late 1980s gene targeting in murine embryonic stem cells (ESCs) enabled the transmission of mutations into the mouse germ line, and emerged as a novel option to study the genetic basis of regulatory networks as they exist in the genome. Still, classical gene targeting proved to be limited in several ways as gene functions became irreversibly destroyed by the marker gene that had to be introduced for selecting recombinant ESCs. These early steps led to animals in which the mutation was present in all cells of the body from the beginning leading to complex phenotypes and/or early lethality. There was a clear need for methods to restrict these mutations to specific points in development and specific cell types. This dream became reality when groups in the USA were able to introduce bacteriophage and yeast-derived site ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P1 Phage
P1 is a temperate bacteriophage that infects ''Escherichia coli'' and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages (e.g. the lambda phage) that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
Lambda phage (coliphage λ, scientific name ''Lambdavirus lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a Temperate (virology), temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infections, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the cytopla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integrase
Retroviral integrase (IN) is an enzyme An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ... produced by a retrovirus (such as HIV) that integrates (forms covalent links between) its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases ( recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination. The macromolecular complex of an IN macromolecule bound to the ends of the viral DNA ends has been referred to as the '' intasome''; IN is a key component in this and the retroviral pre-integration complex. Structure All retroviral IN proteins contain three canonical domains, connected by flexible linkers: * an N-terminal HH-CC zinc-binding domain (a three-heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups ( alpha- , beta- , gamma- amino acids, etc.); other categories relate to polarity, ionization, and side-chain group type ( aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino-acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence. Amino acids are formally named by the IUPAC- IUBMB Joint Commi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminal End
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic acid, carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in Writing system#Directionality, LTR writing systems). This correlates the translation (biology), translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one anot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminal End
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Joe Z
Joe or JOE may refer to: Arts Film and television * ''Joe'' (1970 film), starring Peter Boyle * ''Joe'' (2013 film), starring Nicolas Cage, based on the novel ''Joe'' (1991) by Larry Brown * Joe (2023 film), an Indian film * ''Joe'' (TV series), a British TV series airing from 1966 to 1971 * ''Joe'', a 2002 Canadian animated short about Joe Fortes Music and radio * "Joe" (Inspiral Carpets song) * "Joe" (Red Hot Chili Peppers song) * "Joe", a song by The Cranberries on their album ''To the Faithful Departed'' *"Joe", a song by PJ Harvey on her album '' Dry'' *"Joe", a song by AJR on their album ''OK Orchestra'' * Joe FM (other), any of several radio stations Computing * Joe's Own Editor, a text editor for Unix systems * Joe, an object-oriented Java computing framework based on Sun's Distributed Objects Everywhere project Media * Joe (website), a news website for the UK and Ireland * ''Joe'' (magazine), a defunct periodical developed originally for Kenyan youth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA. DNA ligase is used in both DNA repair and DNA replication (see '' Mammalian ligases''). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see '' Research applications''). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA. Enzymatic mechanism The mechanism of DNA ligase is to form two covalent phosphodiester bonds between ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |