|
Junk DNA
Junk DNA (non-functional DNA) is a DNA sequence that has no known biological function. Most organisms have some junk DNA in their genomes—mostly pseudogenes and fragments of transposons and viruses—but it is possible that some organisms have substantial amounts of junk DNA. All protein-coding regions are generally considered to be functional elements in genomes. Additionally, non-protein coding regions such as genes for ribosomal RNA and transfer RNA, regulatory sequences, origins of replication, centromeres, telomeres, and scaffold attachment regions are considered as functional elements. (See Non-coding DNA for more information.) It is difficult to determine whether other regions of the genome are functional or nonfunctional. There is considerable controversy over which criteria should be used to identify function. Many scientists have an evolutionary view of the genome and they prefer criteria based on whether DNA sequences are preserved by natural selection. Oth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Roy John Britten
Roy John Britten (1 October 1919 – 21 January 2012) was an American molecular biologist known for his discovery of repeated DNA sequences in the genomes of eukaryotic organisms, and later on the evolution of the genome. Early life and education Roy Britten was born in Washington, D.C. He attended Upper Canada College in Toronto, Ontario, and then went to the University of Virginia to study physics. He enrolled at Johns Hopkins University as a graduate student in physics in 1940. At the beginning of World War II, he was recruited to work on the Manhattan Project. In 1951, he received his Ph.D. from Princeton University. His Ph.D. thesis was entitled ''The Scattering of 32 MeV Protons from Several Elements''. Scientific career From 1951 to 1971, he was a staff member at the Carnegie Institution of Washington, Department of Terrestrial Magnetism. While there he attended the phage course at the Cold Spring Harbor Laboratory and started working on the processes by which genetic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposable Element
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome. The discovery of mobile genetic elements earned Barbara McClintock a Nobel Prize in 1983. There are at least two classes of TEs: Class I TEs or retrotransposons generally function via reverse transcription, while Class II TEs or DNA transposons encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. Discovery by Barbara McClintock Barbara McClintock discovered the first TEs in maize (''Zea mays'') at the Cold Spring Harbor Laboratory in New York. McClintock was experimenting with maize plants that had broken chromosomes. In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (style) of the flower received pollen from its own anther. These kernels came from a long line ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Variable Number Tandem Repeat
A variable number tandem repeat (or VNTR) is a location in a genome where a short nucleotide sequence is organized as a tandem repeat. These can be found on many chromosomes, and often show variations in length (number of repeats) among individuals. Each variant acts as an inherited allele, allowing them to be used for personal or parental identification. Their analysis is useful in genetics and biology research, forensics, and DNA fingerprinting. Structure and allelic variation In the schematic above, the rectangular blocks represent each of the repeated DNA sequences at a particular VNTR location. The repeats are in tandem – i.e. they are clustered together and oriented in the same direction. Individual repeats can be removed from (or added to) the VNTR via recombination or replication errors, leading to alleles with different numbers of repeats. Flanking regions are segments of repetitive sequence (shown here as thin lines), allowing the VNTR blocks to be extracte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Long Interspersed Nuclear Element
Long interspersed nuclear elements (LINEs) (also known as long interspersed nucleotide elements or long interspersed elements) are a group of non-LTR (long terminal repeat) retrotransposons that are widespread in the genome of many eukaryotes. LINEs contain an internal RNA polymerase, Pol II promoter to initiate Transcription (biology), transcription into Messenger RNA, mRNA, and encode one or two proteins, ORF1 and ORF2. The functional domains present within ORF1 vary greatly among LINEs, but often exhibit RNA/DNA binding activity. ORF2 is essential to successful retrotransposition, and encodes a protein with both reverse transcriptase and endonuclease activity. LINEs are the most abundant transposable element within the human genome, with approximately 20.7% of the sequences identified as being derived from LINEs. The only active lineage of LINE found within humans belongs to the LINE1, LINE-1 class, and is referred to as L1Hs. The human genome contains an estimated 100,000 tru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alu Elements
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements in the human genome, present in excess of one million copies. Most ''Alu'' elements are thought to be selfish or parasitic DNA. However, it has been suggested that at least some are likely to play a role in evolution and have been used as genetic markers. They are derived from the small cytoplasmic 7SL RNA, a component of the signal recognition particle. ''Alu'' elements are not highly conserved within primate genomes, as only a minority have retained activity, and originated in the genome of an ancestor of Supraprimates. ''Alu'' insertions have been implicated in several inherited human diseases and in various forms of cancer. The study of Alu elements has also been important in elucidating human population genetics and the evolution of primates, including the human evolution, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Short Interspersed Nuclear Element
Short interspersed nuclear elements (SINEs) are non-autonomous, non-coding transposable elements (TEs) that are about 100 to 700 base pairs in length. They are a class of retrotransposons, DNA elements that amplify themselves throughout eukaryotic genomes, often through RNA intermediates. SINEs compose about 13% of the mammalian genome. The internal regions of SINEs originate from tRNA and remain highly conserved, suggesting positive pressure to preserve structure and function of SINEs. While SINEs are present in many species of vertebrates and invertebrates, SINEs are often lineage specific, making them useful markers of divergent evolution between species. Copy number variation and mutations in the SINE sequence make it possible to construct phylogenies based on differences in SINEs between species. SINEs are also implicated in certain types of genetic disease in humans and other eukaryotes. In essence, short interspersed nuclear elements are genetic parasites which have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Retrotransposon
Retrotransposons (also called Class I transposable elements) are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged. Through reverse transcription, retrotransposons amplify themselves quickly to become abundant in eukaryotic genomes such as maize (49–78%) and humans (42%). They are only present in eukaryotes but share features with retroviruses such as HIV, for example, discontinuous reverse transcriptase-mediated extrachromosomal recombination. There are two main types of retrotransposons, long terminal repeats (LTRs) and non-long terminal repeats (non-LTRs). Retrotransposons are classified based on sequence and method of transposition. Most retrotransposons in the maize genome are LTR, whereas in humans they are mostly non-L ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Genome
Bacterial genomes are generally smaller and less variant in size among species when compared with genomes of eukaryotes. Bacterial genomes can range in size anywhere from about 130 kbp to over 14 Mbp. A study that included, but was not limited to, 478 bacterial genomes, concluded that as genome size increases, the number of genes increases at a disproportionately slower rate in eukaryotes than in non-eukaryotes. Thus, the proportion of non-coding DNA goes up with genome size more quickly in non-bacteria than in bacteria. This is consistent with the fact that most eukaryotic nuclear DNA is non-gene coding, while the majority of prokaryotic, viral, and organellar genes are coding. Right now, we have genome sequences from 50 different bacterial phyla and 11 different archaeal phyla. Second-generation sequencing has yielded many draft genomes (close to 90% of bacterial genomes in GenBank are currently not complete); third-generation sequencing might eventually yield a complete genome i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims "to build a comprehensive parts list of functional elements in the human genome." ENCODE also supports further biomedical research by "generating community resources of genomics data, software, tools and methods for genomics data analysis, and products resulting from data analyses and interpretations." The current phase of ENCODE (2016-2019) is adding depth to its resources by growing the number of cell types, data types, assays and now includes support for examination of the mouse genome. History ENCODE was launched by the US National Human Genome Research Institute (NHGRI) in September 2003. Intended as a follow-up to the Human Genome Project, the ENCODE project aims to identify all functional elements in the human genome. The project involves a worldwide consortium of research groups, and data generated from this project can be accessed through public databases. The initial release of ENCODE ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase. There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency. Any piece of DNA with the point centromere DNA sequence on it will typically form a centr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
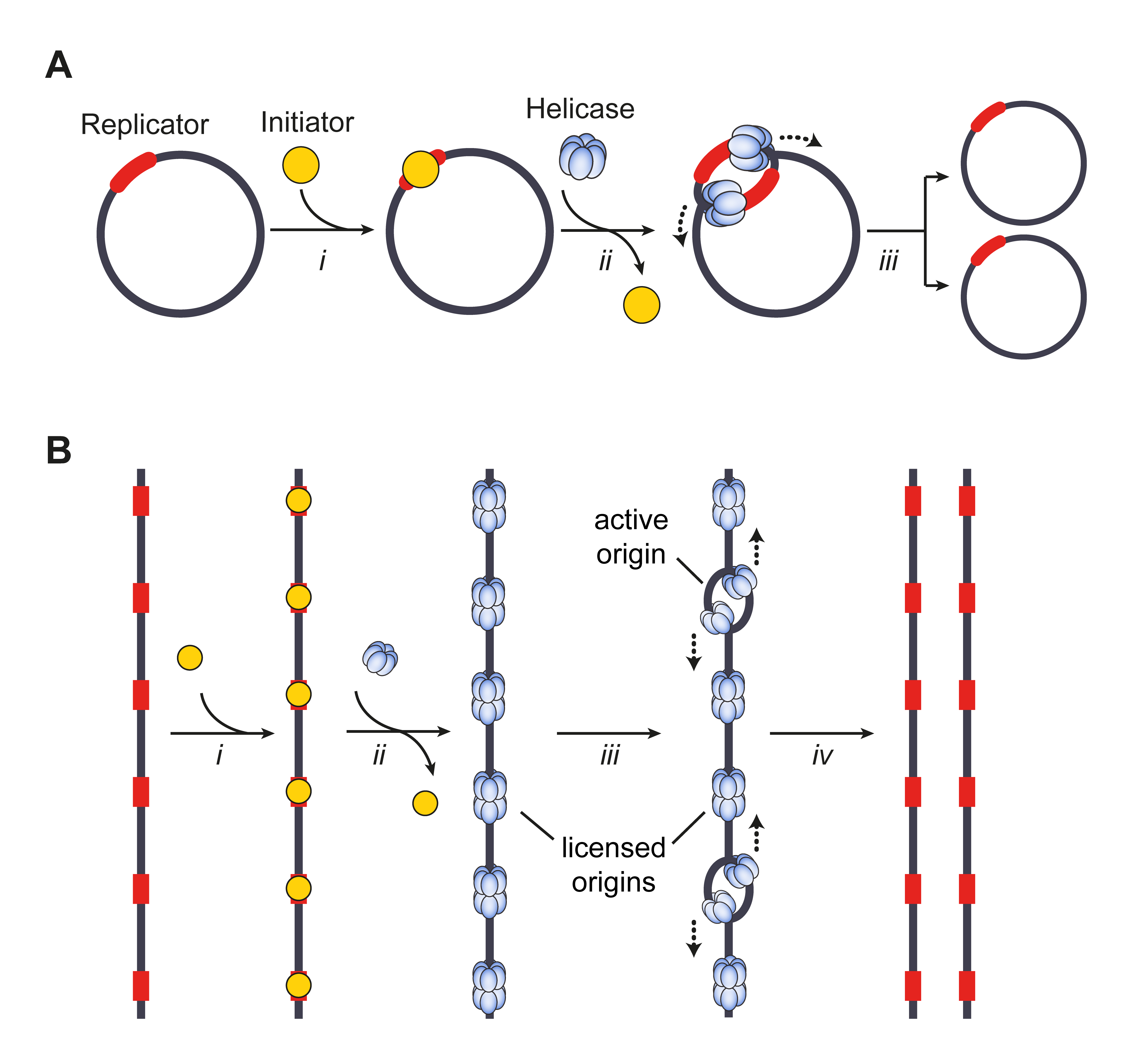
Origin Of Replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. Material was copied from this source, which is available under Creative Commons Attribution 4.0 International License This can either involve the DNA replication, replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA virus, DNA or RNA virus, RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |