|
GyrB
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA polymerase, RNA-polymerase or by helicase in front of the progressing DNA replication#Replication fork, replication fork. The enzyme causes negative DNA supercoil, supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite ''Plasmodium falciparum'' and in chloroplasts of several p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
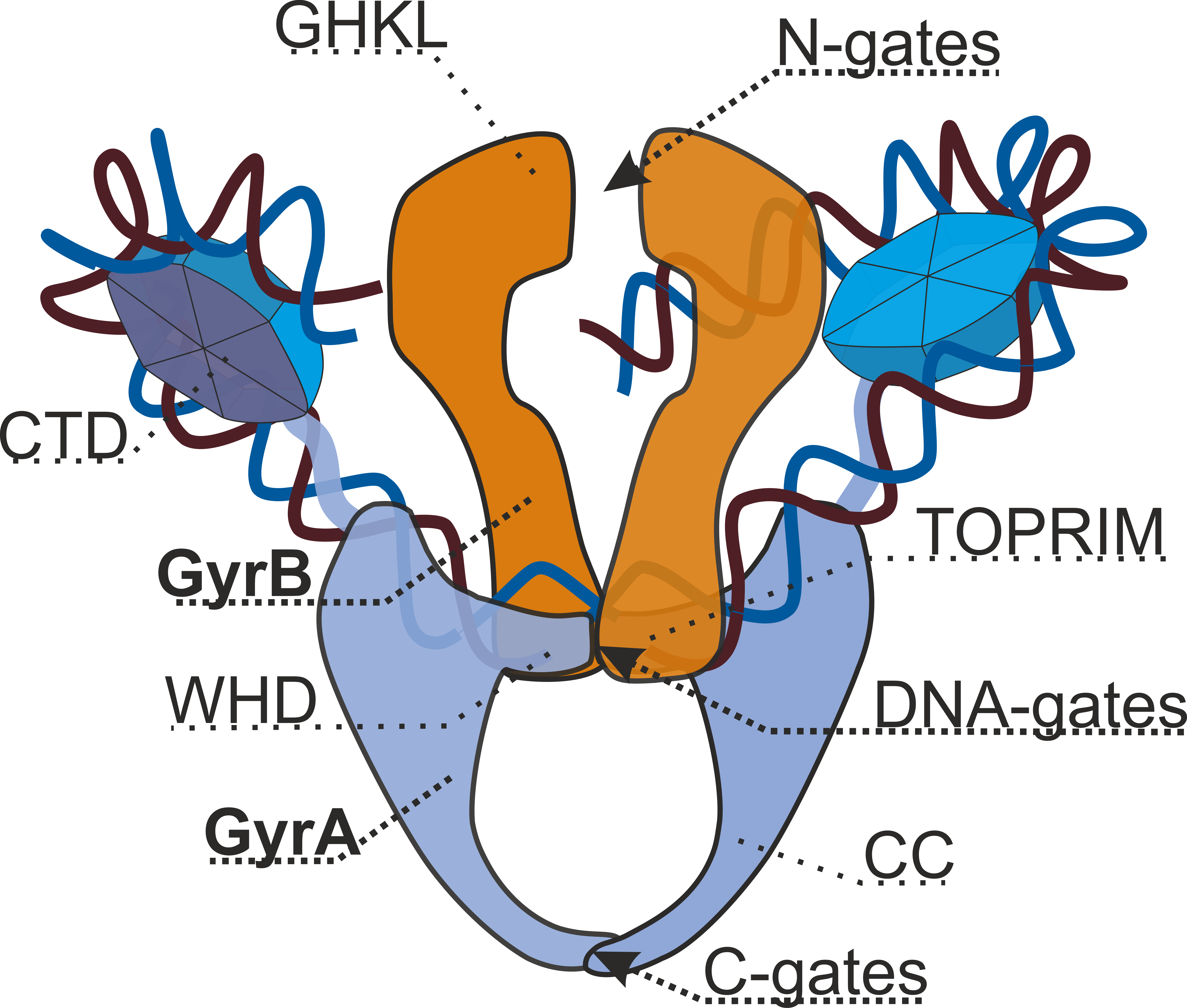
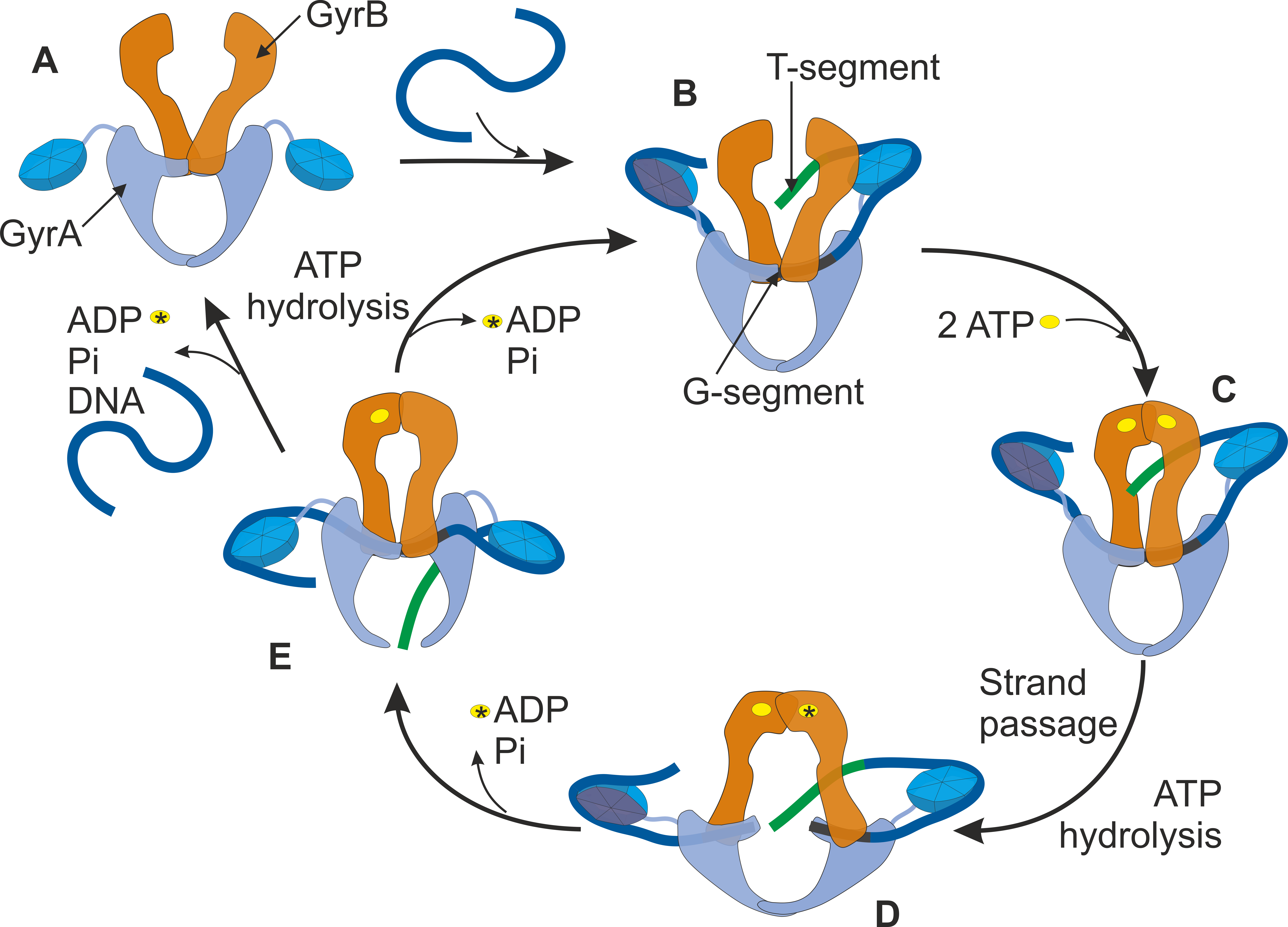
Gyrase Structure Dmitry Sutormin Eng
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite ''Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Novobiocin
Novobiocin, also known as albamycin or cathomycin, is an aminocoumarin antibiotic that is produced by the actinomycete '' Streptomyces niveus'', which has recently been identified as a subjective synonym for ''S. spheroides'' a member of the class Actinomycetia. Other aminocoumarin antibiotics include clorobiocin and coumermycin A1.Alessandra da Silva Eustáquio (2004) Biosynthesis of aminocoumarin antibiotics in Streptomyces: Generation of structural analogues by genetic engineering and insights into the regulation of antibiotic production. DISSERTATION Novobiocin was first reported in the mid-1950s (then called streptonivicin). Clinical use It is active against ''Staphylococcus epidermidis'' and may be used to differentiate it from the other coagulase-negative ''Staphylococcus saprophyticus'', which is resistant to novobiocin, in culture. Novobiocin was licensed for clinical use under the tradename Albamycin (Upjohn) in the 1960s. Its efficacy has been demonstrated in precli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coumermycin A1
Coumermycin A1 is an aminocoumarin. Its main target is the ATPase site of the DNA Gyrase GyrB subunit . See also Chemically induced dimerization References Antibiotics Imidazoles {{antibiotic-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Type II Topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms. In animals, topoisomerase II is a chemotherapy target. In prokaryotes, gyrase is an antibacterial target. Indeed, these enzymes are of interest for a wide range of effects. Function Type II topoisomerases increase or decrease the linking number of a DNA loop by 2 units, and it promotes chromosome disentanglement. For example, DNA gyrase, a type II topoisomerase observed in '' E. coli'' and most other prokaryotes, introduces negative supercoils and decreases the linking number by 2. Gyrase is also able to remove knots from the bacterial chromosome. Along with gyrase, most prokaryotes also contain a second type IIA topoisomerase, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction : deoxynucleoside triphosphate + DNAn pyrophosphate + DNAn+1. DNA polymerase adds nucleotides to the three prime (3')-end of a DNA strand, one nucleotide at a time. Every time a cell divides, DNA polymerases are required to duplicate the cell's DNA, so that a copy of the original DNA molecule can be passed to each daughter cell. In this way, genetic information is passed down from generation to generation. Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminocoumarin
Aminocoumarin is a class of antibiotics that act by an inhibition of the DNA gyrase enzyme involved in the cell division in bacteria. They are derived from ''Streptomyces '' species, whose best-known representative – ''Streptomyces coelicolor'' – was completely sequenced in 2002. The aminocoumarin antibiotics include: * Novobiocin, Albamycin (Pharmacia And Upjohn) * Coumermycin * Clorobiocin Structure The core of aminocoumarin antibiotics is made up of a 3-amino-4,7-dihydroxycumarin ring, which is linked, e.g., with a sugar in 7-Position and a benzoic acid derivative in 3-Position. Clorobiocin is a natural antibiotic isolated from several ''Streptomyces'' strains and differs from novobiocin in that the methyl group at the 8 position in the coumarin ring of novobiocin is replaced by a chlorine atom, and the carbamoyl at the 3' position of the noviose sugar is substituted by a 5-methyl-2-pyrrolylcarbonyl group. Mechanism of action The aminocoumarin antibiotics are known ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibiotics
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the common cold or influenza; drugs which inhibit viruses are termed antiviral drugs or antivirals rather than antibiotics. Sometimes, the term ''antibiotic''—literally "opposing life", from the Greek roots ἀντι ''anti'', "against" and βίος ''bios'', "life"—is broadly used to refer to any substance used against microbes, but in the usual medical usage, antibiotics (such as penicillin) are those produced naturally (by one microorganism fighting another), whereas non-antibiotic antibacterials (such as sulfonamides and antise ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PBR322
pBR322 is a plasmid and was one of the first widely used '' E. coli'' cloning vectors. Created in 1977 in the laboratory of Herbert Boyer at the University of California, San Francisco, it was named after Francisco Bolivar Zapata, the postdoctoral researcher and Raymond L. Rodriguez. The p stands for "plasmid," and BR for "Bolivar" and "Rodriguez." pBR322 is 4361 base pairs in length and has two antibiotic resistance genes – the gene '' bla'' encoding the ampicillin resistance (AmpR) protein, and the gene ''tetA'' encoding the tetracycline resistance (TetR) protein. It contains the origin of replication of pMB1, and the '' rop'' gene, which encodes a restrictor of plasmid copy number. The plasmid has unique restriction sites for more than forty restriction enzymes. Eleven of these forty sites lie within the TetR gene. There are two sites for restriction enzymes HindIII and ClaI within the promoter of the TetR gene. There are six key restriction sites inside the AmpR gene.T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PSC101
pSC101 is a DNA plasmid that is used as a cloning vector in genetic cloning experiments. pSC101 was the first cloning vector, used in 1973 by Herbert Boyer and Stanley Norman Cohen. Using this plasmid they have demonstrated that a gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ... from a frog could be transferred into bacterial cells and then expressed by the bacterial cells. The plasmid is a natural plasmid from ''Salmonella typhimurium''. History In the early 1970s, Herbert Boyer and Stanley Norman Cohen produced pSC101, the first plasmid vector for cloning purposes. Soon after successfully cloning two pSC101 plasmids together to create one large plasmid, they published the results describing the experiment, in 1973. The cloning of genes into plasmids occurred soon af ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage Mu
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus (the first of its kind to be identified) of the family ''Myoviridae'' which has been shown to cause genetic transposition. It is of particular importance as its discovery in ''Escherichia coli'' by Larry Taylor was among the first observations of insertion elements in a genome. This discovery opened up the world to an investigation of transposable elements and their effects on a wide variety of organisms. While Mu was specifically involved in several distinct areas of research (including ''E. coli'', maize, and HIV), the wider implications of transposition and insertion transformed the entire field of genetics. Anatomy Phage Mu is nonenveloped, with a head and a tail. The head has an icosahedral structure of about 54 nm in width. The neck is knob-like, and the tail is contractile with a base plate and six short terminal fibers. The genome has been fully sequenced and consists of 36,717 nucleotides, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |