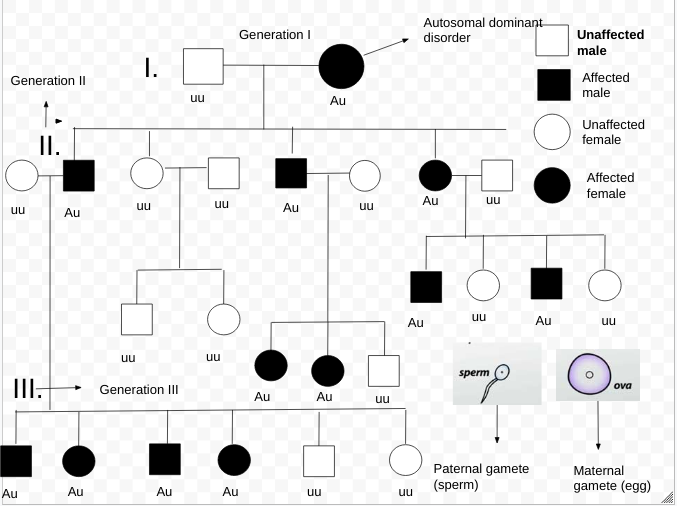
|
Gene Order
Gene order is the permutation of genome arrangement. A fair amount of research has been done trying to determine whether gene orders evolve according to a molecular clock ( molecular clock hypothesis) or in jumps (punctuated equilibrium). By comparing gene orders in dissimilar organisms, scientists are able to develop a molecular phylogeny tree. When organisms have similar gene orders, meaning they have likely diverged recently, it is called synteny. Some research on gene orders in animals' mitochondrial genomes reveals that the mutation rate of gene orders is not a constant in some degrees. Methods for genome mapping, determining the gene order, include: * Radiation hybrid mapping * Genetic linkage mapping (recombinant frequency) * Fluorescent in situ hybridization (FISH) * Restriction mapping * Sequence-tagged site content mapping * Southern hybridization * PCR * Whole genome sequencing Whole genome sequencing (WGS), also known as full genome sequencing or just gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Clock Hypothesis
The molecular clock is a figurative term for a technique that uses the mutation rate of biomolecules to deduce the time in prehistory when two or more life forms diverged. The biomolecular data used for such calculations are usually nucleotide sequences for DNA, RNA, or amino acid sequences for proteins. Early discovery and genetic equidistance The notion of the existence of a so-called "molecular clock" was first attributed to Émile Zuckerkandl and Linus Pauling who, in 1962, noticed that the number of amino acid differences in hemoglobin between different lineages changes roughly linearly with time, as estimated from fossil evidence. They generalized this observation to assert that the rate of evolutionary change of any specified protein was approximately constant over time and over different lineages (known as the molecular clock hypothesis). The genetic equidistance phenomenon was first noted in 1963 by Emanuel Margoliash, who wrote: "It appears that the number of resi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Punctuated Equilibrium
In evolutionary biology, punctuated equilibrium (also called punctuated equilibria) is a Scientific theory, theory that proposes that once a species appears in the fossil record, the population will become stable, showing little evolution, evolutionary change for most of its geological history. : ''Reprinted in'' * * This state of little or no morphological change is called ''stasis''. When significant evolutionary change occurs, the theory proposes that it is generally restricted to rare and geologic time scale, geologically rapid events of branching speciation called cladogenesis. Cladogenesis is the process by which a species splits into two distinct species, rather than one species gradually transforming into another. Punctuated equilibrium is commonly contrasted with phyletic gradualism, the idea that evolution generally occurs uniformly by the steady and gradual transformation of whole lineages (anagenesis). In 1972, paleontologists Niles Eldredge and Stephen Jay Goul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Phylogenetics
Molecular phylogenetics () is the branch of phylogeny that analyzes genetic, hereditary molecular differences, predominantly in DNA sequences, to gain information on an organism's evolutionary relationships. From these analyses, it is possible to determine the processes by which diversity among species has been achieved. The result of a molecular phylogenetics, phylogenetic analysis is expressed in a phylogenetic tree. Molecular phylogenetics is one aspect of molecular systematics, a broader term that also includes the use of molecular data in Taxonomy (biology), taxonomy and biogeography. Molecular phylogenetics and molecular evolution correlate. Molecular evolution is the process of selective changes (mutations) at a molecular level (genes, proteins, etc.) throughout various branches in the tree of life (evolution). Molecular phylogenetics makes inferences of the evolutionary relationships that arise due to molecular evolution and results in the construction of a phylogenetic tre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synteny
In genetics, the term synteny refers to two related concepts: * In classical genetics, ''synteny'' describes the physical co-localization of genetic loci on the same chromosome within an individual or species. * In current biology, ''synteny'' more commonly refers to ''colinearity'', i.e. conservation of blocks of order within two sets of chromosomes that are being compared with each other. These blocks are referred to as ''syntenic blocks''. The Encyclopædia Britannica gives the following description of synteny, using the modern definition: Etymology ''Synteny'' is a neologism meaning "on the same ribbon"; Greek: ', ''syn'' "along with" + ', ''tainiā'' "band". This can be interpreted classically as "on the same chromosome", or in the modern sense of having the same order of genes on two (homologous) strings of DNA (or chromosomes). : co-localization on a chromosome The classical concept is related to genetic linkage: Linkage between two loci is established by the observat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Mapping
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene. The essence of all genome mapping is to place a collection of molecular markers onto their respective positions on the genome. Molecular markers come in all forms. Genes can be viewed as one special type of genetic markers in the construction of genome maps, and mapped the same way as any other markers. In some areas of study, gene mapping contributes to the creation of new recombinants within an organism. Gene maps help describe the spatial arrangement of genes on a chromosome. Genes are designated to a specific location on a chromosome known as the locus and can be used as molecular markers to find the distance between other genes on a chromosome. Maps provide researchers with the opportunity to predict the inheritance patterns of specific traits, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Radiation Hybrid Mapping
Radiation hybrid mapping (also known as RH mapping) is a technique for mapping mammalian chromosomes. Radiation hybrid mapping consists of several steps. A ''radiation hybrid'' is "a cell or organism that contains fragments of chromosomes from a second organism". Radiation hybrids are generated by using X-rays An X-ray (also known in many languages as Röntgen radiation) is a form of high-energy electromagnetic radiation with a wavelength shorter than those of ultraviolet rays and longer than those of gamma rays. Roughly, X-rays have a wavelength ran ... to randomly break chromosomes into fragments, then implanting the fragments into non-irradiated rodent cells, which replicate and thus clone the chromosomes. Then these clones are analyzed for the presence of certain DNA markers. If two given DNA markers are far apart on the initial chromosome, then it is likely that they will appear in distinct fragments. The frequency of the separation of the markers into different fragme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Linkage
Genetic linkage is the tendency of Nucleic acid sequence, DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two Genetic marker, genetic markers that are physically near to each other are unlikely to be separated onto different Chromatid, chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two Gene, genes are on a chromosome, the lower the chance of Genetic recombination, recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets (mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Map
A restriction map is a map of known restriction sites within a sequence of DNA. Restriction mapping requires the use of restriction enzymes, which cleave the DNA at or near the restriction site. In molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by transduction. One approach in constructing a restriction map of a DNA molecule is to sequence the whole molecule and to run the sequence through a computer program that will find the recognition sites that are present for every restriction enzyme known. Before sequencing was automated, it would have been prohibitively expensive to sequence In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence-tagged Site
A sequence-tagged site (or STS) is a short (200 to 500 base pair) DNA sequence that has a single occurrence in the genome and whose location and base sequence are known. Usage STSs can be easily detected by the polymerase chain reaction (PCR) using specific primers. For this reason they are useful for constructing genetic and physical maps from sequence data reported from many different laboratories. They serve as landmarks on the developing physical map of a genome. When STS loci contain genetic polymorphisms (e.g. simple sequence length polymorphisms, SSLPs, single nucleotide polymorphisms), they become valuable genetic markers, i.e. loci which can be used to distinguish individuals. They are used in shotgun sequencing, specifically to aid sequence assembly. STSs are very helpful for detecting microdeletions in some genes. For example, some STSs can be used in screening by PCR to detect microdeletions in Azoospermia (AZF) genes in infertile In biology, infertility is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |