|
Facultative Anaerobes
A facultative anaerobic organism is an organism that makes ATP by aerobic respiration if oxygen is present, but is capable of switching to fermentation if oxygen is absent. Some examples of facultatively anaerobic bacteria are ''Staphylococcus'' spp., ''Escherichia coli'', ''Salmonella'', ''Listeria'' spp., '' Shewanella oneidensis'' and '' Yersinia pestis''. Certain eukaryotes are also facultative anaerobes, including pupfish, fungi such as ''Saccharomyces cerevisiae'' and many aquatic invertebrates such as nereid polychaetes. It has been observed that in mutants of '' Salmonella typhimurium'' that underwent mutations to be either obligate aerobes or anaerobes, there were varying levels of chromatin-remodeling proteins. The obligate aerobes were later found to have a defective DNA gyrase subunit A gene ('' gyrA''), while obligate anaerobes were defective in topoisomerase I (''topI''). This indicates that topoisomerase I and its associated relaxation of chromosomal DNA is requ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anaerobic
Anaerobic means "living, active, occurring, or existing in the absence of free oxygen", as opposed to aerobic which means "living, active, or occurring only in the presence of oxygen." Anaerobic may also refer to: *Adhesive#Anaerobic, Anaerobic adhesive, a bonding agent that does not cure in the presence of air *Anaerobic respiration, respiration in the absence of oxygen, using some other molecule as the final electron acceptor **Anaerobic organism, any organism whose redox metabolism does not depend on free oxygen **Anammox, anaerobic ammonium oxidation, a globally important microbial process of the nitrogen cycle **Anaerobic filter, an anaerobic digester with a tank containing a filter medium where anaerobic microbes can establish themselves **Anaerobic digestion, the use of anaerobic bacteria to break down waste, with biogas as a byproduct ***Anaerobic clarigester, an anaerobic digester that treats dilute biodegradable feedstocks and allows different retention times for solids an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Invertebrates
Invertebrates are animals that neither develop nor retain a vertebral column (commonly known as a ''spine'' or ''backbone''), which evolved from the notochord. It is a paraphyletic grouping including all animals excluding the chordate subphylum Vertebrata, i.e. vertebrates. Well-known phyla of invertebrates include arthropods, molluscs, annelids, echinoderms, flatworms, cnidarians, and sponges. The majority of animal species are invertebrates; one estimate puts the figure at 97%. Many invertebrate taxa have a greater number and diversity of species than the entire subphylum of Vertebrata. Invertebrates vary widely in size, from 10 μm (0.0004 in) myxozoans to the 9–10 m (30–33 ft) colossal squid. Some so-called invertebrates, such as the Tunicata and Cephalochordata, are actually sister chordate subphyla to Vertebrata, being more closely related to vertebrates than to other invertebrates. This makes the "invertebrates" paraphyletic, so the term has ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fumarate
Fumaric acid or ''trans''-butenedioic acid is an organic compound with the formula HO2CCH=CHCO2H. A white solid, fumaric acid occurs widely in nature. It has a fruit-like taste and has been used as a food additive. Its E number is E297. The salts and esters are known as fumarates. Fumarate can also refer to the ion (in solution). Fumaric acid is the ''trans'' isomer of butenedioic acid, while maleic acid is the ''cis'' isomer. Biosynthesis and occurrence It is produced in eukaryotic organisms from succinate in complex 2 of the electron transport chain via the enzyme succinate dehydrogenase. Fumaric acid is found in fumitory (''Fumaria officinalis''), bolete mushrooms (specifically ''Boletus fomentarius var. pseudo-igniarius''), lichen, and Iceland moss. Fumarate is an intermediate in the citric acid cycle used by cells to produce energy in the form of adenosine triphosphate (ATP) from food. It is formed by the oxidation of succinate by the enzyme succinate dehydr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Terminal Oxidases
Terminal may refer to: Computing Hardware * Computer terminal, a set of primary input and output devices for a computer * Terminal (electronics), a device for joining electrical circuits together ** Battery terminal, electrical contact used to connect a load or charger to a single cell or multiple-cell battery * Terminal (telecommunication), a device communicating over a line * Feedback terminal, a physical device used collect anonymous feedback Software * Terminal emulator, a program that emulates a computer terminal within some other display architecture ** Terminal (macOS), a terminal emulator included with macOS ** Windows Terminal, a terminal emulator for Windows 10 and Windows 11 ** GNOME Terminal, a Linux and BSD terminal emulator * Terminal and nonterminal symbols, lexical elements used in specifying the production rules constituting a formal grammar in computer science. Fonts * Terminal (typeface), a monospace font * Terminal (typography), a type of stroke ending ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
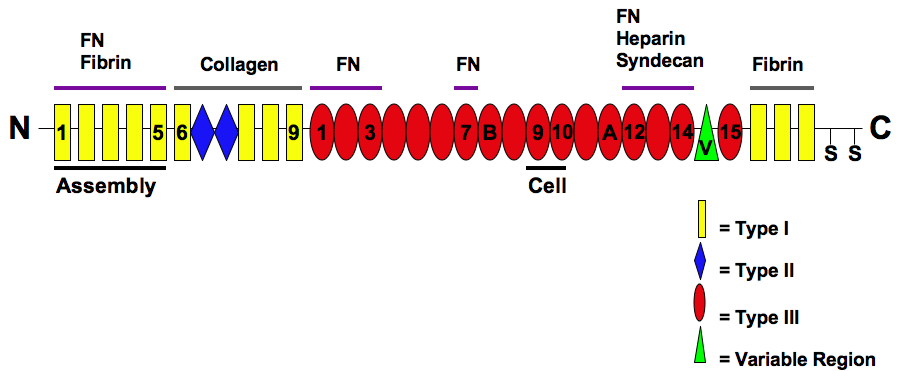
Fibronectin
Fibronectin is a high- molecular weight (~500-~600 kDa) glycoprotein of the extracellular matrix that binds to membrane-spanning receptor proteins called integrins. Fibronectin also binds to other extracellular matrix proteins such as collagen, fibrin, and heparan sulfate proteoglycans (e.g. syndecans). Fibronectin exists as a protein dimer, consisting of two nearly identical monomers linked by a pair of disulfide bonds. The fibronectin protein is produced from a single gene, but alternative splicing of its pre-mRNA leads to the creation of several isoforms. Two types of fibronectin are present in vertebrates: * soluble plasma fibronectin (formerly called "cold-insoluble globulin", or CIg) is a major protein component of blood plasma (300 μg/ml) and is produced in the liver by hepatocytes. * insoluble cellular fibronectin is a major component of the extracellular matrix. It is secreted by various cells, primarily fibroblasts, as a soluble protein dimer and is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are Gene expression, expressed in the desired Cell (biology), cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Core
Core or cores may refer to: Science and technology * Core (anatomy), everything except the appendages * Core (laboratory), a highly specialized shared research resource * Core (manufacturing), used in casting and molding * Core (optical fiber), the signal-carrying portion of an optical fiber * Core, the central part of a fruit * Hydrophobic core, the interior zone of a protein * Nuclear reactor core, a portion containing the fuel components * Pit (nuclear weapon) or core, the fissile material in a nuclear weapon * Semiconductor intellectual property core (IP core), is a unit of design in ASIC/FPGA electronics and IC manufacturing * Atomic core, an atom with no valence electrons * Lithic core, in archaeology, a stone artifact left over from toolmaking Geology and astrophysics * Core sample, in Earth science, a sample obtained by coring ** Ice core * Core, the central part of a galaxy; see Mass deficit * Core (anticline), the central part of an anticline or syncline * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pasteur Effect
The Pasteur effect describes how available oxygen inhibits ethanol fermentation, driving yeast to switch toward aerobic respiration for increased generation of the energy carrier adenosine triphosphate (ATP). More generally, in the medical literature, the Pasteur effect refers to how the presence of oxygen causes in a decrease in the cellular rate of glycolysis and suppression of lactate accumulation. The effect occurs in animal tissues, as well as in microorganisms belonging to the fungal kingdom. Discovery In 1857, microbiologist Louis Pasteur showed that aeration of yeasted broth causes cell growth to increase while the fermentation rate decreases, based on lowered ethanol production. Explanation Yeast fungi, being facultative anaerobes, can either produce energy through ethanol fermentation or aerobic respiration. When the O2 concentration is low, the two pyruvate molecules formed through glycolysis are each fermented into ethanol and carbon dioxide. While only 2 ATP ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphofructokinase
Phosphofructokinase (PFK) is a kinase enzyme that phosphorylates fructose 6-phosphate in glycolysis. Function The enzyme-catalysed transfer of a phosphoryl group from ATP is an important reaction in a wide variety of biological processes. Phosphofructokinase catalyses the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate, a key regulatory step in the glycolytic pathway. It is allosterically inhibited by ATP and allosterically activated by AMP, thus indicating the cell's energetic needs when it undergoes the glycolytic pathway. PFK exists as a homotetramer in bacteria and mammals (where each monomer possesses 2 similar domains) and as an octomer in yeast (where there are 4 alpha- (PFK1) and 4 beta-chains (PFK2), the latter, like the mammalian monomers, possessing 2 similar domains). This protein may use the morpheein model of allosteric regulation. PFK is about 300 amino acids in length, and structural studies of the bacterial enzyme have show ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TOP1
DNA topoisomerase 1 is an enzyme that in humans is encoded by the ''TOP1'' gene. It is a DNA topoisomerase, an enzyme that catalyzes the transient breaking and rejoining of a single strand of DNA. Function This gene encodes a DNA topoisomerase, an enzyme that controls and alters the topologic states of DNA during transcription. This enzyme catalyzes the transient breaking and rejoining of a single strand of DNA which lets the broken strand rotate around the intact strand, thus altering the topology of DNA. This gene is localized to chromosome 20 and has pseudogenes which reside on chromosomes 1 and 22. Mechanism As reviewed by Champoux, the type IB topoisomerases, including TOP1, form a covalent intermediate in which the active site tyrosine becomes attached to the 3' phosphate end of the cleaved strand rather than the 5' phosphate end. The eukaryotic topoisomerases I were found to nick the DNA with a preference for a sequence of nucleotides that extends from positions -4 t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
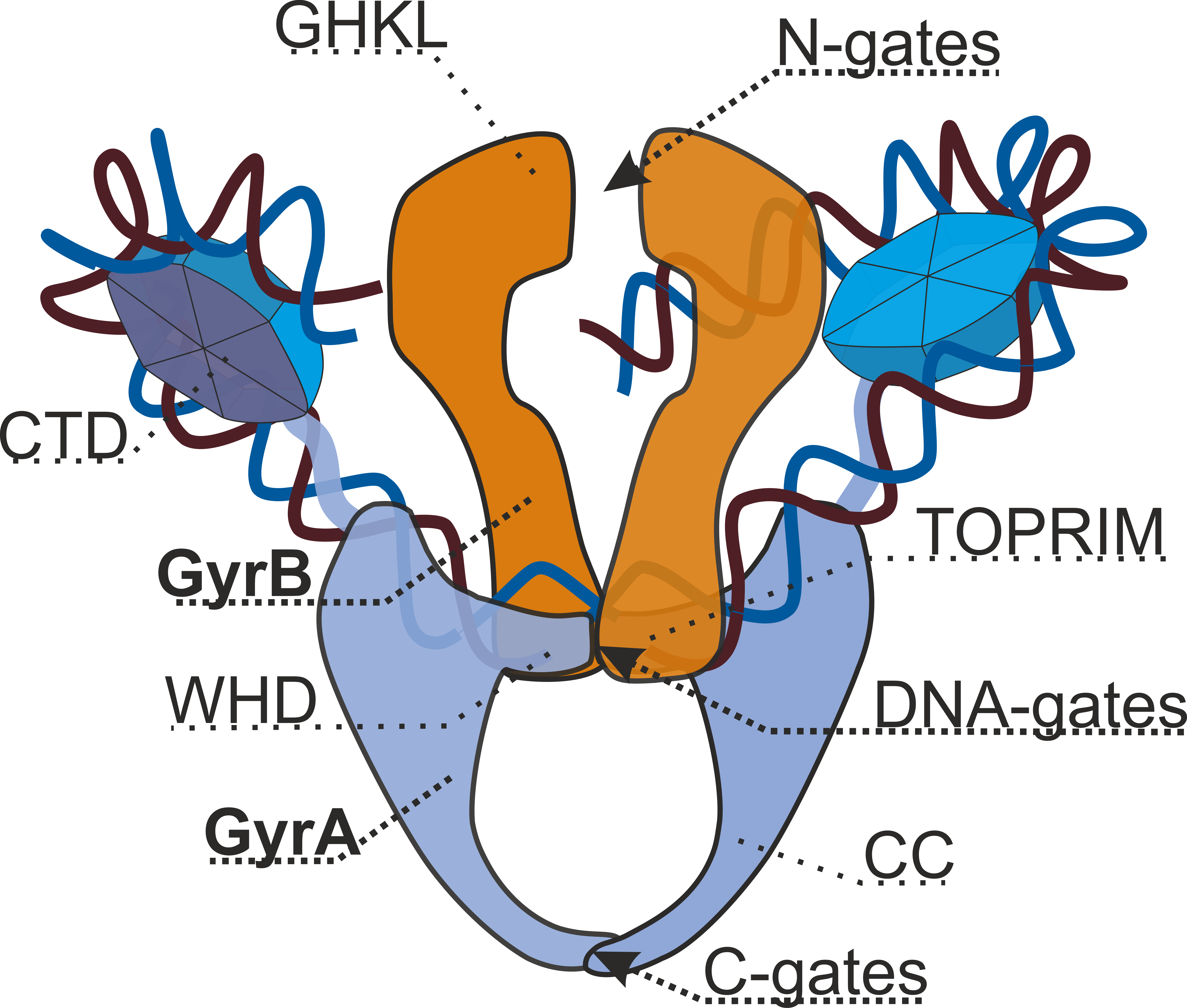
DNA Gyrase
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite ''Plasmodium falciparum'' and in chloroplasts of several plants. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |