|
Diguanylate Cyclase
In enzymology, diguanylate cyclase, also known as diguanylate kinase (), is an enzyme that catalysis, catalyzes the chemical reaction: 2 Guanosine triphosphate ↔ 2 diphosphate + cyclic di-3',5'-guanylate The Substrate (biochemistry), substrates of diguanylate cyclases (DGCs) are two molecules of guanosine triphosphate (GTP) and the product (biochemistry), products are two molecules of diphosphate and one molecule of cyclic di-3’,5’-guanylate (cyclic di-GMP). Degradation of cyclic di-GMP to guanosine monophosphate (GMP) is catalyzed by a phosphodiesterase (PDE). Structure Diguanylate cyclases are characterized by the conserved amino acid sequence motifs “GGDEF domain, GGDEF” (Gly-Gly-Aspartic acid, Asp-Glutamic acid, Glu-Phenylalanine, Phe) or “GGEEF” (Gly-Gly-Glu-Glu-Phe), which constitute the protein domain, domain of the DGC active site. These domains are often found coupled to other signaling domains within multidomain proteins. Often, GGDEF domains with DGC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-di-GMP
Cyclic di-GMP (also called cyclic diguanylate and c-di-Guanosine monophosphate, GMP) is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in ''Dictyostelium''. The biological role of cyclic di-GMP was first uncovered when it was identified as an allosteric activator of a cellulose synthase found in ''Gluconacetobacter xylinus'' in order to produce microbial cellulose. In structure, it is a cycle containing only two guanine bases linked by ribose and phosphate. Contact with surfaces increases c-di-GMP which increases gene transcription, transcription, gene translation, translation, and Post-translational modification, post translation of exopolysaccharides (EPSs) and other extracellular polymeric substance matrix components (see the review by Jenal et al 2017). In bacteria, certain signals are communicated by synthesizing or degrading cyclic di-GMP. Cyclic di-GMP i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glutamic Acid
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synthesize enough for its use. It is also the most abundant excitatory neurotransmitter in the vertebrate nervous system. It serves as the precursor for the synthesis of the inhibitory gamma-aminobutyric acid (GABA) in GABA-ergic neurons. Its molecular formula is . Glutamic acid exists in three optically isomeric forms; the dextrorotatory -form is usually obtained by hydrolysis of gluten or from the waste waters of beet-sugar manufacture or by fermentation.Webster's Third New International Dictionary of the English Language Unabridged, Third Edition, 1971. Its molecular structure could be idealized as HOOC−CH()−()2−COOH, with two carboxyl groups −COOH and one amino group −. However, in the solid state and mildly acidic water sol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
β-sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined version wa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crystal Structure
In crystallography, crystal structure is a description of the ordered arrangement of atoms, ions or molecules in a crystalline material. Ordered structures occur from the intrinsic nature of the constituent particles to form symmetric patterns that repeat along the principal directions of three-dimensional space in matter. The smallest group of particles in the material that constitutes this repeating pattern is the unit cell of the structure. The unit cell completely reflects the symmetry and structure of the entire crystal, which is built up by repetitive translation of the unit cell along its principal axes. The translation vectors define the nodes of the Bravais lattice. The lengths of the principal axes, or edges, of the unit cell and the angles between them are the lattice constants, also called ''lattice parameters'' or ''cell parameters''. The symmetry properties of the crystal are described by the concept of space groups. All possible symmetric arrangements of partic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenylate Cyclase
Adenylate cyclase (EC 4.6.1.1, also commonly known as adenyl cyclase and adenylyl cyclase, abbreviated AC) is an enzyme with systematic name ATP diphosphate-lyase (cyclizing; 3′,5′-cyclic-AMP-forming). It catalyzes the following reaction: :ATP = 3′,5′-cyclic AMP + diphosphate It has key regulatory roles in essentially all cells. It is the most polyphyletic known enzyme: six distinct classes have been described, all catalyzing the same reaction but representing unrelated gene families with no known sequence or structural homology. The best known class of adenylyl cyclases is class III or AC-III (Roman numerals are used for classes). AC-III occurs widely in eukaryotes and has important roles in many human tissues. All classes of adenylyl cyclase catalyse the conversion of adenosine triphosphate (ATP) to 3',5'-cyclic AMP (cAMP) and pyrophosphate.Magnesium ions are generally required and appear to be closely involved in the enzymatic mechanism. The cAMP produced by A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally-occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
A dimer () (''di-'', "two" + ''-mer'', "parts") is an oligomer consisting of two monomers joined by bonds that can be either strong or weak, covalent or intermolecular. Dimers also have significant implications in polymer chemistry, inorganic chemistry, and biochemistry. The term ''homodimer'' is used when the two molecules are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Under special conditions, most OH-containing molecules form dimers, e.g. the water dimer. Excimers and exciplexes are excited structures with a short life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain isobutyl group, making it a non-polar aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, and beans and other legumes. It is encoded by the codons UUA, UUG, CUU, CUC, CUA, and CUG. Like valine and isoleucine, leucine is a branched-chain amino acid. The primary metabolic end products of leucine metabolism are acetyl-CoA and acetoacetate; consequently, it is one of the two exclusively ketogenic amino acids, with lysine being the other. It is the most imp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
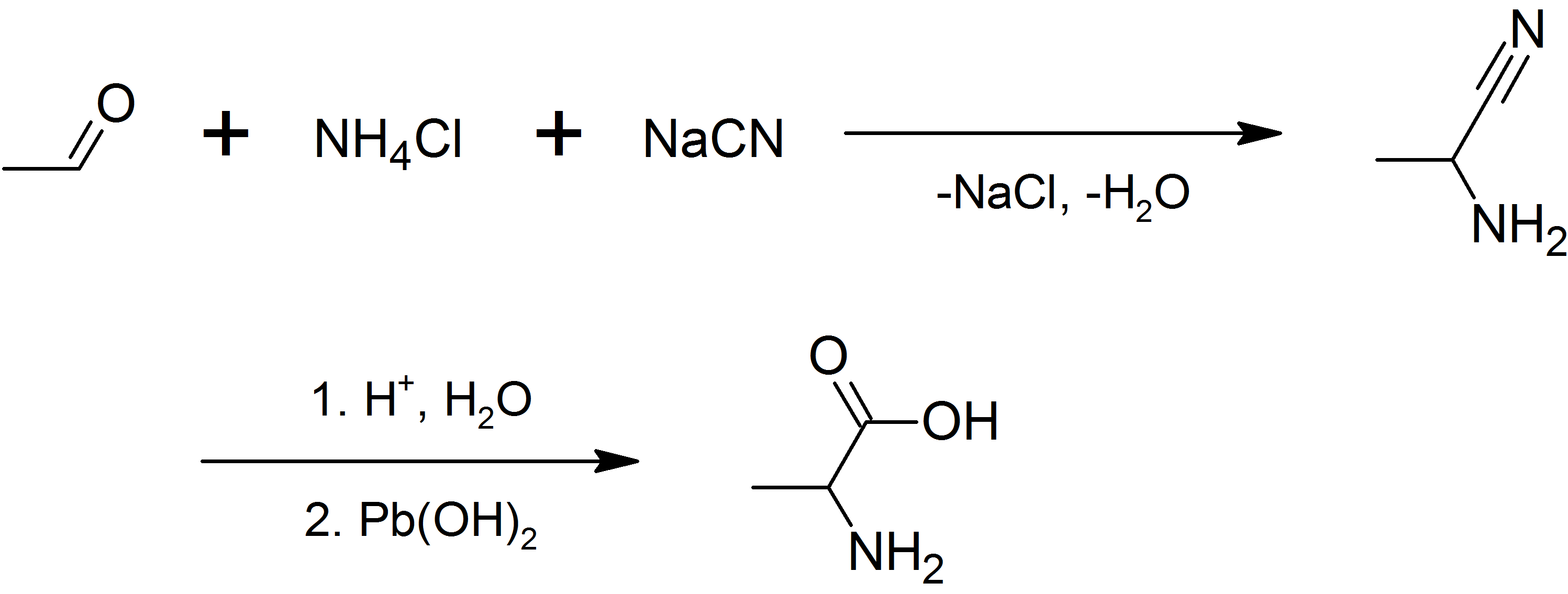
Alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently, its IUPAC systematic name is 2-aminopropanoic acid, and it is classified as a nonpolar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated (as −NH3+) and its carboxyl group deprotonated (as −CO2−). It is non-essential to humans as it can be synthesised metabolically and does not need to be present in the diet. It is encoded by all codons starting with GC (GCU, GCC, GCA, and GCG). The L-isomer of alanine (left-handed) is the one that is incorporated into proteins. L-alanine is second only to leucine in rate of occurrence, accounting for 7.8% of the primary structure in a sample of 1,150 proteins. The right-handed form, D-alanine, occurs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EAL Domain
In molecular biology, the EAL domain is a conserved protein domain. It is found in diverse bacterial signalling proteins. It is named EAL after its conserved residues. The EAL domain may function as a diguanylate phosphodiesterase A phosphodiesterase (PDE) is an enzyme that breaks a phosphodiester bond. Usually, ''phosphodiesterase'' refers to cyclic nucleotide phosphodiesterases, which have great clinical significance and are described below. However, there are many o .... The domain contains many conserved acidic residues that could participate in metal binding and might form the phosphodiesterase active site. References {{InterPro content, IPR001633 Protein domains ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |