|
Cyclic Di-GMP
Cyclic di-GMP (also called cyclic diguanylate and c-di- GMP) is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in ''Dictyostelium''. The biological role of cyclic di-GMP was first uncovered when it was identified as an allosteric activator of a cellulose synthase found in '' Gluconacetobacter xylinus'' in order to produce microbial cellulose. In structure, it is a cycle containing only two guanine bases linked by ribose and phosphate. Function Contact with surfaces increases c-di-GMP which increases transcription, translation, and post translation of exopolysaccharides (EPSs) and other extracellular polymeric substance matrix components (see the review by Jenal et al 2017). In bacteria, certain signals are communicated by synthesizing or degrading cyclic di-GMP. Cyclic di-GMP is synthesized by proteins with diguanylate cyclase activity. These proteins t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanosine Monophosphate
Guanosine monophosphate (GMP), also known as 5′-guanidylic acid or guanylic acid (conjugate base guanylate), is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside guanosine. GMP consists of the phosphate group, the pentose sugar ribose, and the nucleobase guanine; hence it is a ribonucleotide monophosphate. Guanosine monophosphate is commercially produced by microbial fermentation. As an acyl substituent, it takes the form of the prefix guanylyl-. ''De novo'' synthesis GMP synthesis starts with D-ribose 5′-phosphate, a product of the pentose phosphate pathway. The synthesis proceeds by the gradual formation of the purine ring on carbon-1 of ribose, with CO2, glutamine, glycine, aspartate and one-carbon derivatives of tetrahydrofolate donating various elements towards the building of the ring. As inhibitor of guanosine monophosphate synthesis in experimental models, the glutamine analogue DON can be used.''Ahluwalia GS et al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diguanylate Cyclase
In enzymology, diguanylate cyclase, also known as diguanylate kinase (), is an enzyme that catalyzes the chemical reaction: 2 Guanosine triphosphate ↔ 2 diphosphate + cyclic di-3',5'-guanylate The substrates of diguanylate cyclases (DGCs) are two molecules of guanosine triphosphate (GTP) and the products are two molecules of diphosphate and one molecule of cyclic di-3’,5’-guanylate (cyclic di-GMP). Degradation of cyclic di-GMP to guanosine monophosphate (GMP) is catalyzed by a phosphodiesterase (PDE). Structure Diguanylate cyclases are characterized by the conserved amino acid sequence motifs “ GGDEF” ( Gly-Gly- Asp- Glu- Phe) or “GGEEF” (Gly-Gly-Glu-Glu-Phe), which constitute the domain of the DGC active site. These domains are often found coupled to other signaling domains within multidomain proteins. Often, GGDEF domains with DGC activity are found in the same proteins as c-di-GMP-specific phosphodiesterase (PDE) EAL (Glu- Ala- Leu) domains. DGC is thou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
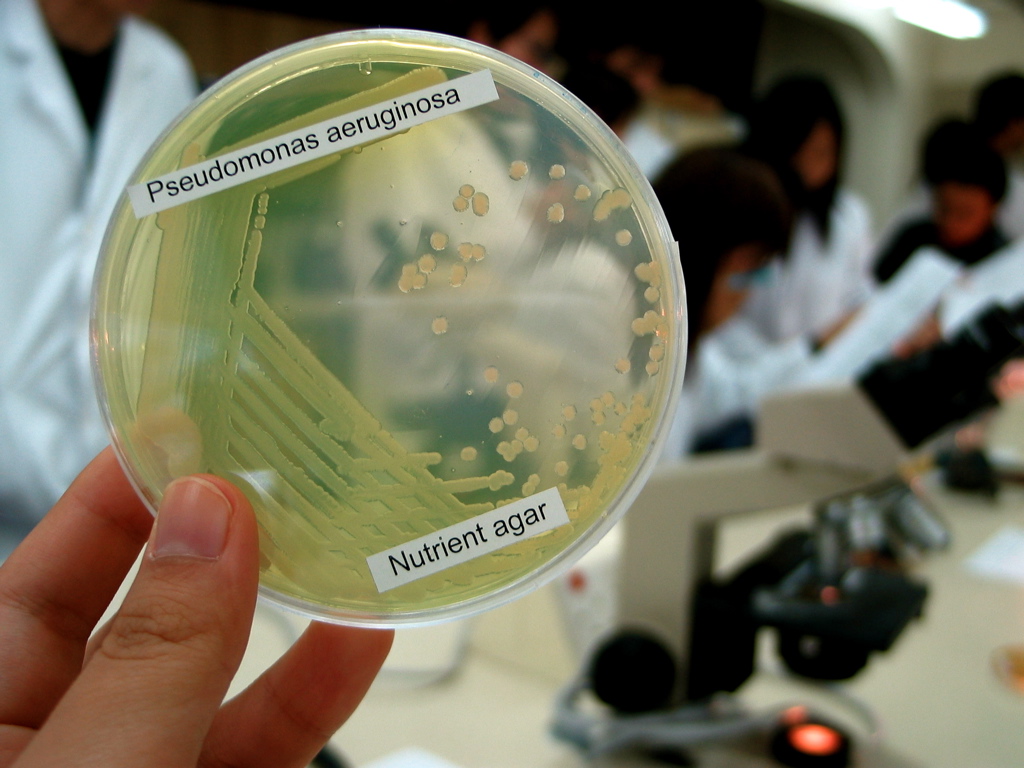
Pseudomonas Aeruginosa
''Pseudomonas aeruginosa'' is a common Bacterial capsule, encapsulated, Gram-negative bacteria, Gram-negative, Aerobic organism, aerobic–facultative anaerobe, facultatively anaerobic, Bacillus (shape), rod-shaped bacteria, bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, ''P. aeruginosa'' is a multiple drug resistance, multidrug resistant pathogen recognized for its ubiquity, its Intrinsic and extrinsic properties, intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes. ''P. aeruginosa'' is able to selectively inhibit various antibiotics from penetrating its outer membrane'' ''– and has high resistance to several antibiotics. According to the World Health Organization ''P. aeruginosa'' poses one of the greatest threats to humans in terms of an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caulobacter Crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935). ''C. crescentus'' is an important model organism for studying the regulation of the cell cycle, asymmetric cell division, and cellular differentiation. ''Caulobacter'' daughter cells have two very different forms. One daughter is a mobile "swarmer" cell that has a single flagellum at one cell pole that provides swimming motility for chemotaxis. The other daughter, called the "stalked" cell, has a tubular stalk structure protruding from one pole that has an adhesive holdfast material on its end, with which the stalked cell can adhere to surfaces. Swarmer cells differentiate into stalked cells after a short period of motility. Chromosome replication and cell division only occurs in the stalked cell stage. ''C. crescentus'' derives its name from its crescent shape, wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclic Di-GMP-II Riboswitch
Cyclic di-GMP-II riboswitches (also c-di-GMP-II riboswitches) form a class of riboswitches that specifically bind cyclic di-GMP, a second messenger used in multiple bacterial processes such as virulence, motility and biofilm formation. Cyclic di-GMP II riboswitches are structurally unrelated to cyclic di-GMP-I riboswitches, though they have the same function. Cyclic di-GMP-II riboswitches were discovered by bioinformatics, and are common in species within the class Clostridia and the genus '' Deinococcus''. They are also found in some other bacterial lineages. There is significant overlap between species that use cyclic di-GMP-I and cyclic di-GMP-II riboswitches, as both riboswitch classes are common in Clostridia. In ''Clostridioides difficile (bacteria)'' strains, a cyclic di-GMP-II riboswitch is found adjacent to a group I catalytic intron. Group I introns are ribozymes that catalyze the splicing of the RNA molecule in which they are embedded. In the riboswitch-associ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclic Di-GMP-I Riboswitch
Cyclic di-GMP-I riboswitches are a class of riboswitch that specifically bind cyclic di-GMP, which is a second messenger that is used in a variety of microbial processes including virulence, motility and biofilm formation. Cyclic di-GMP-I riboswitches were originally identified by bioinformatics as a conserved RNA-like structure called the "GEMM motif". These riboswitches are present in a wide variety of bacteria, and are most common in Clostridia and certain varieties of Pseudomonadota. The riboswitches are present in pathogens such as '' Clostridioides difficile'', ''Vibrio cholerae'' (which causes cholera) and '' Bacillus anthracis'' (which causes anthrax). '' Geobacter uraniumreducens'' is predicted to have 30 instances of this riboswitch in its genome. A bacteriophage that infects ''C. difficile'' is predicted to carry a cyclic di-GMP-I riboswitch, which it might use to detect and exploit the physiological state of bacteria that it infects. The discovery of this ribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Riboswitch
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in Translation (biology), production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its Effector (biology), effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, ribozyme, catalyze reactions, or to bind other RNA or protein macromolecules. The original definition of the term "riboswitch" specified that they directly sense small-molecule metabolite concentrations. Although this definition remains in common use, some biologists have used a broader definition that includes other cis-regulatory element, cis-regulatory RNAs. However, this article will discuss o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAS Domain
A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. Generally, the PAS domain acts as a molecular sensor, whereby small molecules and other proteins associate via binding of the PAS domain. Due to this sensing capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor. Discovery PAS domains are found in a large number of organisms from bacteria to mammals. The PAS domain was named after the three proteins in which it was first discovered: * Per – period circadian protein * Arnt – aryl hydrocarbon receptor nuclear translocator protein * Sim – single-minded protein Since the initial discovery of the PAS domain, a large quantity of PAS domain binding sites have been discovered in bacteria and eukaryotes. A subset called PAS LOV proteins are responsive to oxygen, light ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EAL Domain
In molecular biology, the EAL domain is a conserved protein domain. It is found in diverse bacterial signalling proteins. It is named EAL after its conserved residues. The EAL domain may function as a diguanylate phosphodiesterase. The domain contains many conserved acidic residues that could participate in metal binding and might form the phosphodiesterase active site In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s .... References {{InterPro content, IPR001633 Protein domains ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virulence
Virulence is a pathogen's or microorganism's ability to cause damage to a host. In most cases, especially in animal systems, virulence refers to the degree of damage caused by a microbe to its host. The pathogenicity of an organism—its ability to cause disease—is determined by its virulence factors. In the specific context of gene for gene systems, often in plants, virulence refers to a pathogen's ability to infect a resistant host. Virulence can also be transferred using a plasmid. The noun ''virulence'' (Latin noun ) derives from the adjective ''virulent'', meaning disease severity. The word ''virulent'' derives from the Latin word ''virulentus'', meaning "a poisoned wound" or "full of poison". The term ''virulence'' does not only apply to viruses. From an ecological standpoint, virulence is the loss of fitness induced by a parasite upon its host. Virulence can be understood in terms of proximate causes—those specific traits of the pathogen that help make the host ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Motility
Motility is the ability of an organism to move independently using metabolism, metabolic energy. This biological concept encompasses movement at various levels, from whole organisms to cells and subcellular components. Motility is observed in animals, microorganisms, and even some plant structures, playing crucial roles in activities such as foraging, reproduction, and cellular functions. It is genetically determined but can be influenced by environmental factors. In multicellular organisms, motility is facilitated by systems like the Nervous system, nervous and Human musculoskeletal system, musculoskeletal systems, while at the cellular level, it involves mechanisms such as amoeboid movement and flagellar propulsion. These cellular movements can be directed by external stimuli, a phenomenon known as taxis. Examples include chemotaxis (movement along chemical gradients) and phototaxis (movement in response to light). Motility also includes physiological processes like gastroi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
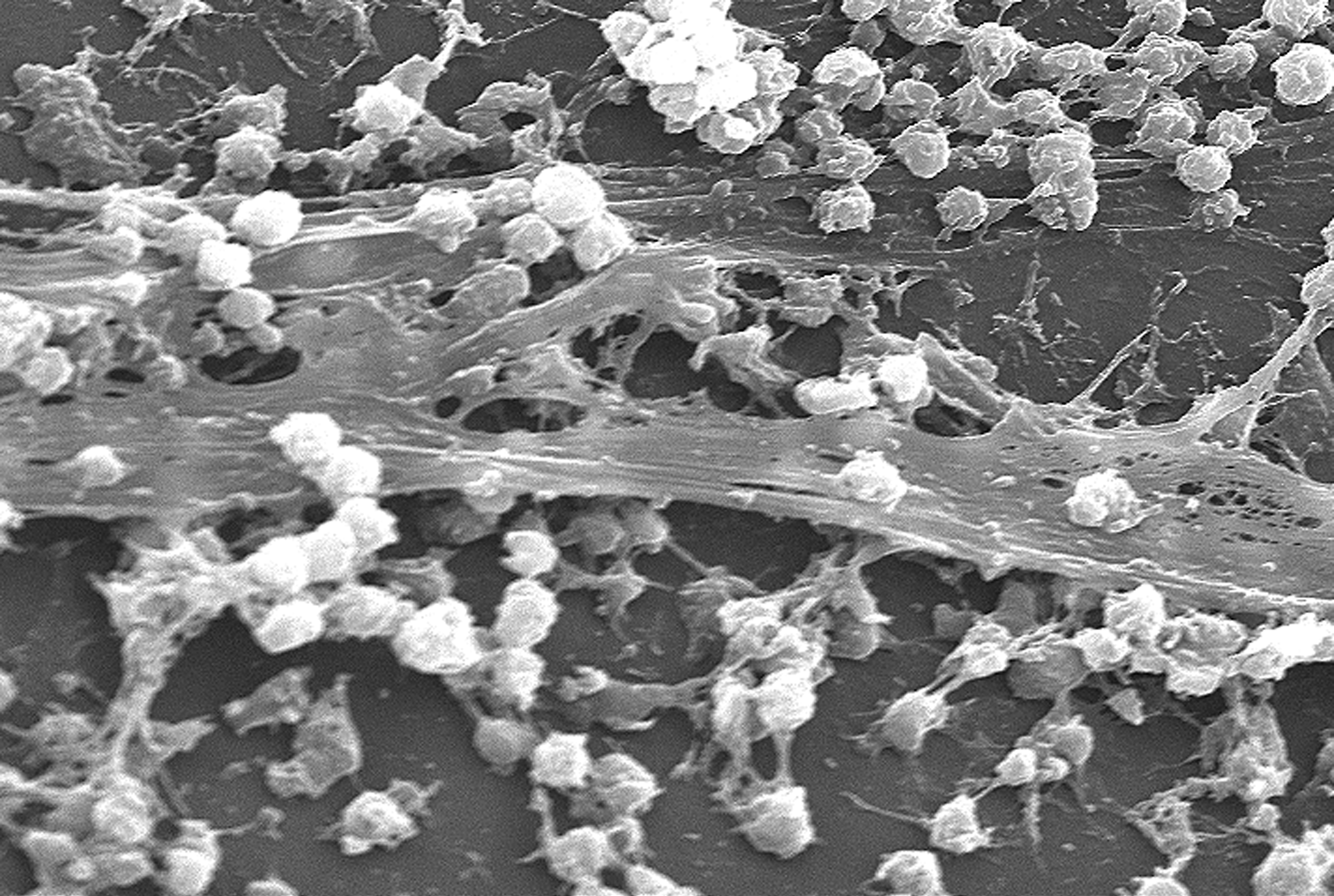
Biofilm
A biofilm is a Syntrophy, syntrophic Microbial consortium, community of microorganisms in which cell (biology), cells cell adhesion, stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric combination of extracellular polysaccharides, proteins, lipids and DNA. Because they have a three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes". Biofilms may form on living (biotic) or non-living (abiotic) surfaces and can be common in natural, industrial, and hospital settings. They may constitute a microbiome or be a portion of it. The microbial cells growing in a biofilm are physiology, physiologically distinct from planktonic cells of the same organism, which, by contrast, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |