|
Chimera (molecular Biology)
In molecular biology, and more importantly high-throughput DNA sequencing, a chimera is a single DNA sequence originating when multiple transcripts or DNA sequences get joined. Chimeras can be considered artifacts and be filtered out from the data during processing to prevent spurious inferences of biological variation. However, chimeras should not be confused with chimeric reads, which are generally used by structural variant callers to detect structural variation events and are not always an indication of the presence of a chimeric transcript or gene. In a different context, the deliberate creation of artificial chimeras can also be a useful tool in molecular biology. For example, in protein engineering, "chimeragenesis" (forming chimeras between proteins that are encoded by homologous cDNAs) p. 424 is one of the "two major techniques used to manipulate cDNA sequences". For gene fusions that occur through natural processes, see chimeric genes and fusion genes. Descriptio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primer (molecular Biology)
A primer is a short, single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer is a type of oligo, short for oligonucleotide. DNA polymerases (responsible for DNA replication) are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replaced with DNA nucleotides. This forms a gap region known as a nick that is filled in using a ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chimeric Gene
Chimeric genes (literally, made of parts from different sources) form through the combination of portions of two or more coding sequences to produce new genes. These mutations are distinct from fusion genes which merge whole gene sequences into a single reading frame and often retain their original functions. Formation Chimeric genes can form through several different means. Many chimeric genes form through errors in DNA replication or DNA repair so that pieces of two different genes are inadvertently combined.Rogers, RL, Bedford, T and Hartl DL"Formation and Longevity of Chimeric and Duplicate Genes in ''Drosphila''" ''Genetics''. 181: 313-322. Chimeric genes can also form through retrotransposition where a retrotransposon accidentally copies the transcript of a gene and inserts it into the genome in a new location. Depending on where the new retrogene appears, it can recruit new exons to produce a chimeric gene. Chimeric genes can also form through Ectopic recombination, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chimera (genetics)
A genetic chimerism or chimera ( or ) is a single organism composed of cells of different genotype, genotypes. Animal chimeras can be produced by the fusion of two (or more) embryos. In plants and some animal chimeras, Mosaic (genetics), mosaicism involves distinct types of tissue that originated from the same zygote but differ due to mutation during ordinary cell division. Normally, genetic chimerism is not visible on casual inspection; however, it has been detected in the course of proving parentage. More practically, in agronomy, "chimera" indicates a plant or portion of a plant whose tissues are made up of two or more types of cells with different genetic makeup; it can derive from a bud mutation or, more rarely, at the grafting point, from the concrescence of cells of the two bionts; in this case it is commonly referred to as a "graft hybrid", although it is not a hybrid in the genetic sense of "hybrid". In contrast, an individual where each cell contains genetic materi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trans-splicing
''Trans''-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archaea also have "half-genes" for tRNAs. Genic ''trans''-splicing Whereas "normal" (''cis''-)splicing processes a single molecule, ''trans''-splicing generates a single RNA transcript from multiple separate pre-mRNAs. This phenomenon can be exploited for molecular therapy to address mutated gene products. Genic trans-splicing allows variability in RNA diversity and increases proteome complexity. Oncogenesis While some fusion transcripts occur via ''trans''-splicing in normal human cells, ''trans''-splicing can also be the mechanism behind certain oncogenic fusion transcripts. SL ''trans''-splicing Spliced leader (SL) ''trans''-splicing is used by certain microorganisms, notably protists of the Kinetoplastea class to express genes. I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transgene
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the phenotype of an organism. ''Transgene'' describes a segment of DNA containing a gene sequence that has been isolated from one organism and is introduced into a different organism. This non-native segment of DNA may either retain the ability to produce RNA or protein in the transgenic organism or alter the normal function of the transgenic organism's genetic code. In general, the DNA is incorporated into the organism's germ line. For example, in higher vertebrates this can be accomplished by injecting the foreign DNA into the cell nucleus, nucleus of a fertilized ovum. This technique is routinely used to introduce human disease genes or other genes of interest into strains of Laboratory mouse, laboratory mice to study the function or pathol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins (). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA (mRNA) chain. Ribosomes bind to the messenger RNA molecules and use the RNA's sequence of nucleotides to determine the sequence of amino acids needed to generate a protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the riboso ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oncostatin M
Oncostatin M, also known as OSM, is a protein that in humans is encoded by the ''OSM'' gene. OSM is a pleiotropic cytokine that belongs to the interleukin 6 group of cytokines. Of these cytokines it most closely resembles leukemia inhibitory factor (LIF) in both structure and function. As yet poorly defined, it is proving important in liver development, haematopoeisis, inflammation and possibly CNS development. It is also associated with bone formation and destruction. * OSM signals through cell surface receptors that contain the protein gp130. The type I receptor is composed of gp130 and LIFR, the type II receptor is composed of gp130 and OSMR. Discovery, isolation and cloning The human form of OSM was originally isolated in 1986 from the growth media of PMA treated U-937 histiocytic lymphoma cells by its ability to inhibit the growth of cell lines established from melanomas and other solid tumours. A robust protein, OSM is stable between pH2 and 11 and resistant to heati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CYP2C18
Cytochrome P450 2C18 is a protein that in humans is encoded by the ''CYP2C18'' gene. Function This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein localizes to the endoplasmic reticulum but its specific substrate has not yet been determined. The gene is located within a cluster of cytochrome P450 genes on chromosome 10q24. An additional gene, CYP2C17, was once thought to exist; however, CYP4217 is now considered an artefact based on a chimera of CYP2C18 and CYP2C19. CYP2C18 also possesses epoxygenase activity: it can attack various long-chain polyunsaturated fatty acids at their double (i.e. alkene) bonds to form epoxide products that act as signaling agents. It metabolizes: 1) arachidonic acid to various epoxyeicosatrienoic acids (also termed EETs); 2) linoleic acid to 9,10-epoxy oct ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C2orf3
GC-rich sequence DNA-binding factor is a protein that in humans is encoded by the ''GCFC2'' gene. The first mRNA transcript isolated for this gene was part of an artificial chimera derived from two distinct gene transcripts and a primer used in the cloning process (see Genbank accession M29204). A positively charged amino terminus present only in the chimera was determined to bind GC-rich DNA, thus mistakenly thought to identify a transcription factor In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ... gene. References External links * Further reading * * * * * * * * {{gene-2-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
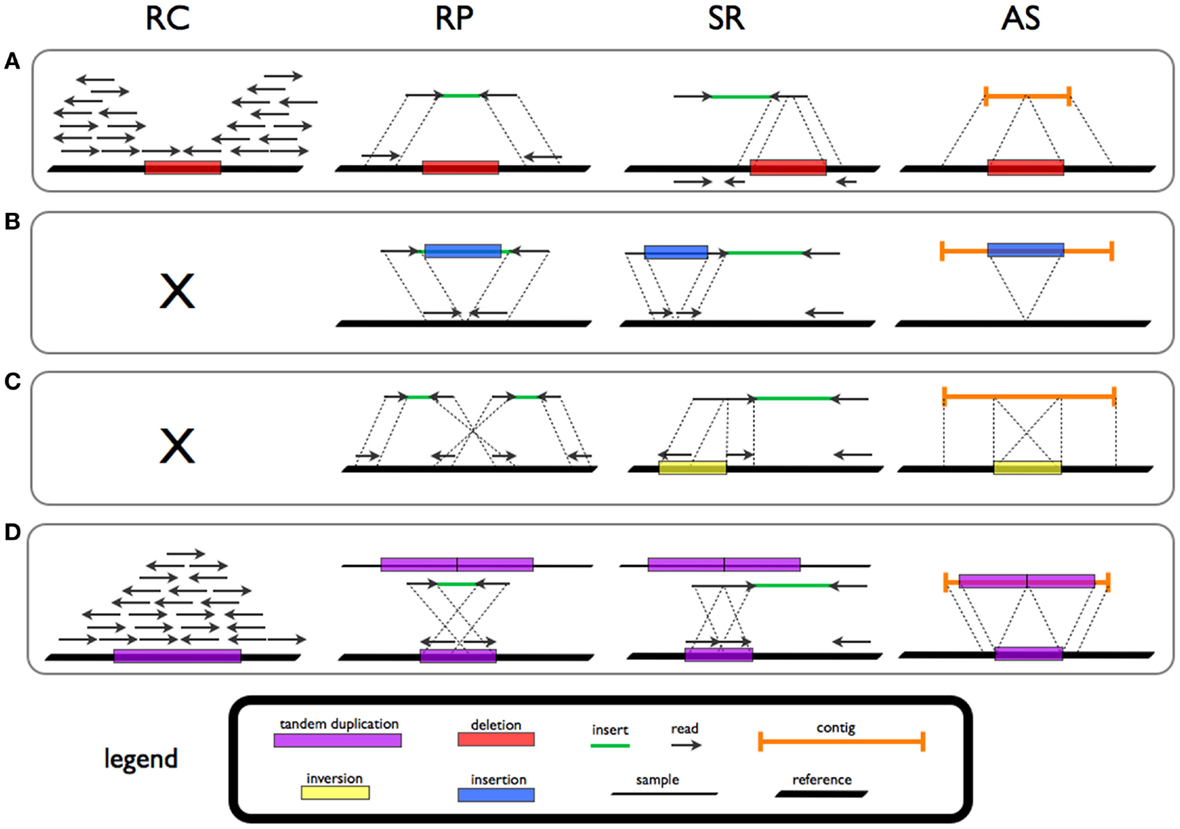
Structural Variation
Genomic structural variation is the variation in structure of an organism's chromosome, such as deletions, duplications, copy-number variants, insertions, inversions and translocations. Originally, a structure variation affects a sequence length about 1kb to 3Mb, which is larger than SNPs and smaller than chromosome abnormality (though the definitions have some overlap). However, the operational range of structural variants has widened to include events > 50bp. Some structural variants are associated with genetic diseases, however most are not. Approximately 13% of the human genome is defined as structurally variant in the normal population, and there are at least 240 genes that exist as homozygous deletion polymorphisms in human populations, suggesting these genes are dispensable in humans. While humans carry a median of 3.6 Mbp in SNPs (compared to a reference genome), a median of 8.9 Mbp is affected by structural variation which thus causes most genetic differences between ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reference Genome
A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the genome, set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead, a reference provides a haploid mosaic of different DNA sequences from each donor. For example, one of the most recent human reference genomes, assembly ''GRCh38, GRCh38/hg38'', is derived from >60 genomic library, genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |