|
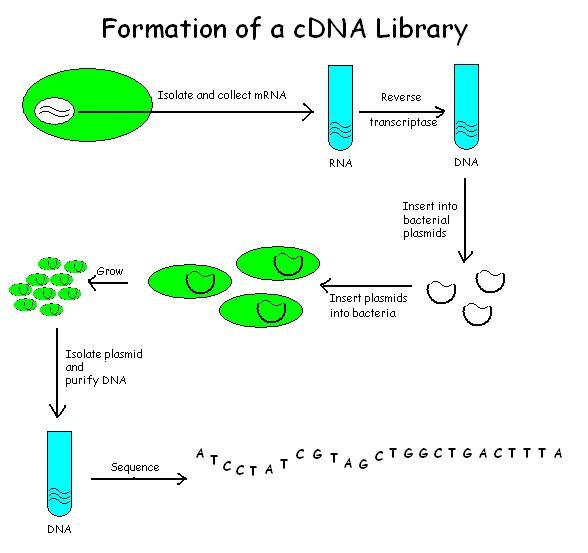
CDNA Library
A cDNA library is a combination of cloned cDNA (complementary DNA) fragments inserted into a collection of host cells, which constitute some portion of the transcriptome of the organism and are stored as a "library". cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue-specific cDNA libraries can be produced. In eukaryotic cells the mature mRNA is already spliced, hence the cDNA produced lacks introns and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library. cDNA Library Construction cDNA is created from a mature mRNA from a eukaryotic cell with the use of reverse transcriptase. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engineered forms. In engineered forms, it often is a copy (replicate) of the naturally occurring DNA from any particular organism's natural genome; the organism's own mRNA was naturally transcribed from its DNA, and the cDNA is reverse transcribed from the mRNA, yielding a duplicate of the original DNA. Engineered cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression as part of recombinant DNA, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trizol
TRIzol is a widely used chemical solution used in the extraction of DNA, RNA, and proteins from cells. The solution was initially used and published by Piotr Chomczyński and Nicoletta Sacchi in 1987. TRIzol is the brand name of guanidinium thiocyanate from the Ambion part of Life Technologies (Thermo Fisher Scientific), Life Technologies, and Tri-Reagent is the brand name from MRC, which was founded by Chomczynski. Uses in extraction The correct name of the method is guanidinium thiocyanate-phenol-chloroform extraction. The use of TRIzol can result in DNA yields comparable to other extraction methods, and it leads to >50% bigger RNA yield. An alternative method for RNA extraction is phenol extraction and TCA/acetone precipitation. Chloroform should be exchanged with 1-bromo-3-chloropropane when using the new generation TRI Reagent. DNA and RNA from TRIzol and TRI reagent can also be extracted using the Direct-zol Miniprep kit by Zymo Research. This method eliminates the use of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomic DNA Library
In molecular biology, a library is a collection of genetic material fragments that are stored and propagated in a population of microbes through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries (formed from reverse-transcribed RNA), genomic libraries (formed from genomic DNA) and randomized mutant libraries (formed by de novo gene synthesis where alternative nucleotides or codons are incorporated). DNA library technology is a mainstay of current molecular biology, genetic engineering, and protein engineering, and the applications of these libraries depend on the source of the original DNA fragments. There are differences in the cloning vectors and techniques used in library preparation, but in general each DNA fragment is uniquely inserted into a cloning vector and the pool of recombinant DNA molecules is then transferred into a population of bacteria (a Bacterial Artificial Chromosome or BAC library) or yeast such that ea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Functional Cloning
Functional cloning is a molecular cloning technique that relies on prior knowledge of the encoded protein’s Protein sequence, sequence or Protein function, function for gene identification. In this assay, a Genomic library, genomic or cDNA library is screened to identify the genetic sequence of a protein of interest. Expression vector, Expression cDNA libraries may be screened with Antibody, antibodies specific for the protein of interest or may rely on selection via the protein function. Historically, the amino acid sequence of a protein was used to prepare degenerate oligonucleotides which were then Hybridization probe, probed against the library to identify the gene encoding the protein of interest. Once candidate clones carrying the gene of interest are identified, they are DNA sequencing, sequenced and their identity is confirmed. This method of cloning allows researchers to screen entire genomes without prior knowledge of the location of the gene or the genetic sequence. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Genetics
Reverse genetics is a method in molecular genetics that is used to help understand the function(s) of a gene by analysing the phenotypic effects caused by genetically engineering specific nucleic acid sequences within the gene. The process proceeds in the opposite direction to forward genetic screens of classical genetics. While forward genetics seeks to find the genetic basis of a phenotype or trait, reverse genetics seeks to find what phenotypes are controlled by particular genetic sequences. Automated DNA sequencing generates large volumes of genomic sequence data relatively rapidly. Many genetic sequences are discovered in advance of other, less easily obtained, biological information. Reverse genetics attempts to connect a given genetic sequence with specific effects on the organism. Reverse genetics systems can also allow the recovery and generation of infectious or defective viruses with desired mutations. This allows the ability to study the virus ''in vitro'' and ''in v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cloning
Cloning is the process of producing individual organisms with identical genomes, either by natural or artificial means. In nature, some organisms produce clones through asexual reproduction; this reproduction of an organism by itself without a mate is known as parthenogenesis. In the field of biotechnology, cloning is the process of creating cloned organisms of Cell (biology), cells and of DNA fragments. The artificial cloning of organisms, sometimes known as reproductive cloning, is often accomplished via somatic-cell nuclear transfer (SCNT), a cloning method in which a viable embryo is created from a somatic cell and an egg cell. In 1996, Dolly (sheep), Dolly the sheep achieved notoriety for being the first mammal cloned from a somatic cell. Another example of artificial cloning is molecular cloning, a technique in molecular biology in which a single living cell is used to clone a large population of cells that contain identical DNA molecules. In bioethics, there are a vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA. DNA ligase is used in both DNA repair and DNA replication (see '' Mammalian ligases''). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see '' Research applications''). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA. Enzymatic mechanism The mechanism of DNA ligase is to form two covalent phosphodiester bonds between ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S1 Nuclease
Nuclease S1 () is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalysis, catalyses the following chemical reaction : Endonucleolytic cleavage to 5'-phosphomononucleotide and 5'-phosphooligonucleotide end-products Although its primary substrate is single-stranded, it can also occasionally introduce single-stranded breaks in double-stranded DNA or RNA, or DNA-RNA hybrids. The enzyme hydrolyses single stranded region in duplex DNA such as loops or gaps. It also cleaves a strand opposite a nick on the complementary strand. It has no sequence specificity. Well-known versions include S1 found in ''Aspergillus oryzae'' (yellow koji mold) and Nuclease P1 found in ''Penicillium citrinum''. Members of the S1/P1 family are found in both prokaryotes and eukaryotes and are thought to be associated in programmed cell death and also in tissue differentiation. Furthermore, they are secretion, secreted extracellular, that is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hairpin Loop
Stem-loops are nucleic acid secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regions of the same nucleic acid strand, usually complementary in nucleotide sequence, base-pair to form a double helix that ends in a loop of unpaired nucleotides. Stem-loops are most commonly found in RNA, and are a key building block of many RNA secondary structures. Stem-loops can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop is dependent on the stability of the helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatche ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalysis, catalyze the chemical reaction : deoxynucleoside triphosphate + DNAn pyrophosphate + DNAn+1. DNA polymerase adds nucleotides to the Directionality (molecular biology), three prime (3')-end of a DNA strand, one nucleotide at a time. Every time a Cell division, cell divides, DNA polymerases are required to duplicate the cell's DNA, so that a copy of the original DNA molecule can be passed to each daughter cell. In this way, genetic information is passed down from generation to generation. Before replication can take place, an enzy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |