|
Biological Age
Biomarkers of aging are biomarkers that could predict functional capacity at some later age better than chronological age. Stated another way, biomarkers of aging would give the true "biological age", which may be different from the chronological age. Validated biomarkers of aging would allow for testing interventions to extend lifespan, because changes in the biomarkers would be observable throughout the lifespan of the organism. Although maximum lifespan would be a means of validating biomarkers of aging, it would not be a practical means for long-lived species such as humans because longitudinal studies would take far too much time. Ideally, biomarkers of aging should assay the biological process of aging and not a predisposition to disease, should cause a minimal amount of trauma to assay in the organism, and should be reproducibly measurable during a short interval compared to the lifespan of the organism. An assemblage of biomarker data for an organism could be termed its "a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic Mechanisms
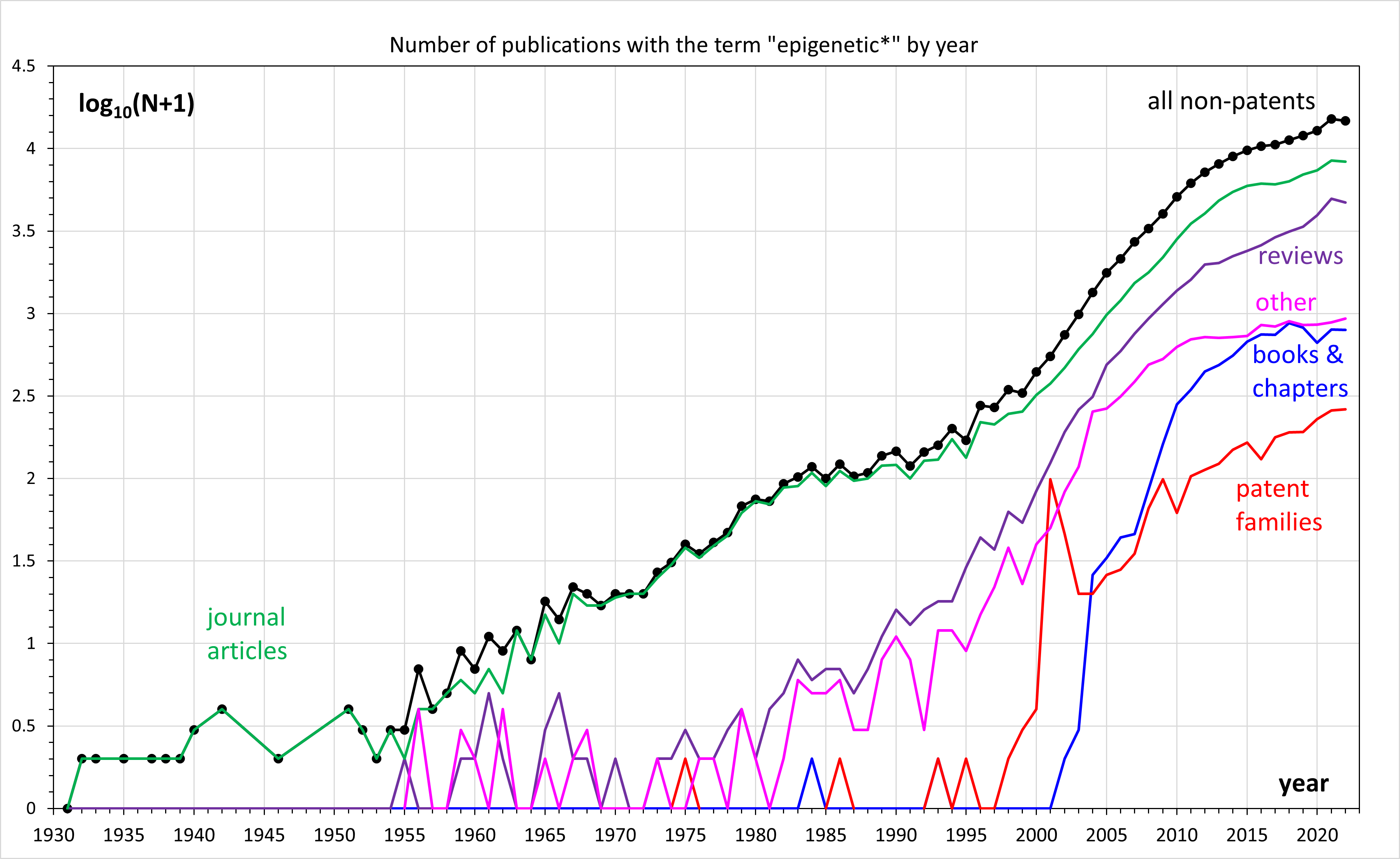
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30- nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 9 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones, which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H4K16ac
H4K16ac is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the acetylation at the 16th lysine residue of the histone H4 protein. H4K16ac is unusual in that it has both transcriptional activation AND repression activities. The loss of H4K20me3 along with a reduction of H4K16ac is a strong indicator of cancer. Lysine acetylation and deacetylation Proteins are typically acetylated on lysine residues and this reaction relies on acetyl-coenzyme A as the acetyl group donor. In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typically, these reactions are catalyzed by enzymes with ''histone acetyltransferase'' (HAT) or ''histone deacetylase'' (HDAC) activity, although HATs and HDACs can modify the acetylation status of non-histone proteins as well. The regulation of transcription factors, effector proteins, molecular ch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
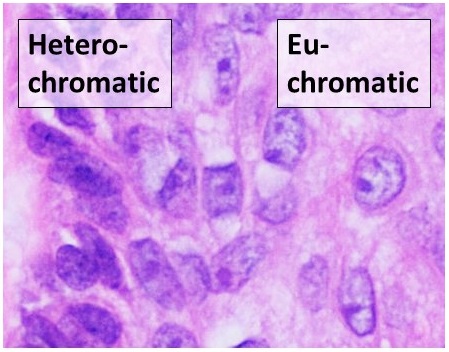
Euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic. In eukaryotes, euchromatin comprises the most active portion of the genome within the cell nucleus. In prokaryotes, euchromatin is the ''only'' form of chromatin present; this indicates that the heterochromatin structure evolved later along with the nucleus, possibly as a mechanism to handle increasing genome size. Structure Euchromatin is composed of repeating subunits known as nucleosomes, reminiscent of an unfolded set of beads on a string, that are approximately 11 nm in diameter. At the core of these nucleosomes are a set of four histone protein pairs: H3, H4, H2A, and H2B. Each core histone protein possesses a 'tail' structu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetylation And Deacetylation
Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminus, N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation. Histone acetylation and deacetylation are essential parts of Regulation of gene expression, gene regulation. These reactions are typically catalysed by enzymes with "histone acetyltransferase" (HAT) or "histone deacetylase" (HDAC) activity. Acetylation is the process where an acetyl functional group is transferred from one molecule (in this case, acetyl coenzyme A) to another. Deacetylation is simply the reverse reaction where an acetyl group is removed from a molecule. Acetylated histones, octameric proteins that organize chromatin into nucleosomes, the basic structural unit of the chromosomes and ultimately higher order structures, represent a type of epigenetic marker within chromatin. Acetylation removes the positive charge on the histones, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are Gene expression, expressed in the desired Cell (biology), cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Senescence
Senescence () or biological aging is the gradual deterioration of Function (biology), functional characteristics in living organisms. Whole organism senescence involves an increase in mortality rate, death rates or a decrease in fecundity with increasing age, at least in the later part of an organism's biological life cycle, life cycle. However, the resulting effects of senescence can be delayed. The 1934 discovery that calorie restriction can Life extension, extend lifespans by 50% in rats, the existence of species having negligible senescence, and the existence of potentially immortal organisms such as members of the genus ''Hydra (genus), Hydra'' have motivated research into Life extension, delaying senescence and thus age-related diseases. Rare human mutations can cause accelerated aging diseases. Environmental Gerontogens, factors may affect aging – for example, overexposure to ultraviolet radiation accelerates skin aging. Different parts of the body may age at different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of DNA, chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery The existence of a special structure at the ends of chromosomes was independently proposed in 1938 by Hermann Joseph Muller, studying the fruit fly ''Drosophila melanogaster'', and in 1939 by Barbara McClintock, working with maize. Muller observed that the ends of irradiated fruit fly chromosomes did not present alterations such as deletions or inversions. He hypothesized the presence of a protective cap, which he coined "telomeres", from the Greek ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Senescence
Cellular senescence is a phenomenon characterized by the cessation of cell division. In their experiments during the early 1960s, Leonard Hayflick and Paul Moorhead found that normal human fetal fibroblasts in culture reach a maximum of approximately 50 cell population doublings before becoming senescent. This process called the Hayflick limit is also known as "replicative senescence", since it is brought about through replication. Hayflick's discovery of mortal cells paved the path for the discovery and understanding of cellular aging molecular pathways. Cellular senescence can be initiated by a wide variety of stress inducing factors. These stress factors include both environmental and internal damaging events, abnormal cellular growth, oxidative stress, autophagy factors, among many other things. The physiological importance for cell senescence has been attributed to prevention of carcinogenesis, and more recently, aging, development, and tissue repair. Senescent cells c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibroblast
A fibroblast is a type of cell (biology), biological cell typically with a spindle shape that synthesizes the extracellular matrix and collagen, produces the structural framework (Stroma (tissue), stroma) for animal Tissue (biology), tissues, and plays a critical role in wound healing. Fibroblasts are the most common cells of connective tissue in animals. Structure Fibroblasts have a branched cytoplasm surrounding an elliptical, speckled cell nucleus, nucleus having two or more nucleoli. Active fibroblasts can be recognized by their abundant rough endoplasmic reticulum (RER). Inactive fibroblasts, called 'fibrocytes', are smaller, spindle-shaped, and have less RER. Although disjointed and scattered when covering large spaces, fibroblasts often locally align in parallel clusters when crowded together. Unlike the epithelial cells lining the body structures, fibroblasts do not form flat monolayers and are not restricted by a polarizing attachment to a basal lamina on one side, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA Box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence. The TATA box is considered a non-coding DNA sequence (also known as a cis-regulatory element). It was termed the "TATA box" as it contains a consensus sequence characterized by repeating T and A base pairs. How the term "box" originated is unclear. In the 1980s, while investigating nucleotide sequences in mouse genome loci, the Hogness box sequence was found and "boxed in" at the -31 position. When consensus nucleotides and alternative ones were compared, homologous regions were "boxed" by the researchers. The boxing in of sequences sheds light on the origin of the term "box". The TATA box was first identified in 1978 as a component of eukaryotic promoters. Transcription is initiated at the TATA box in TA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |