|
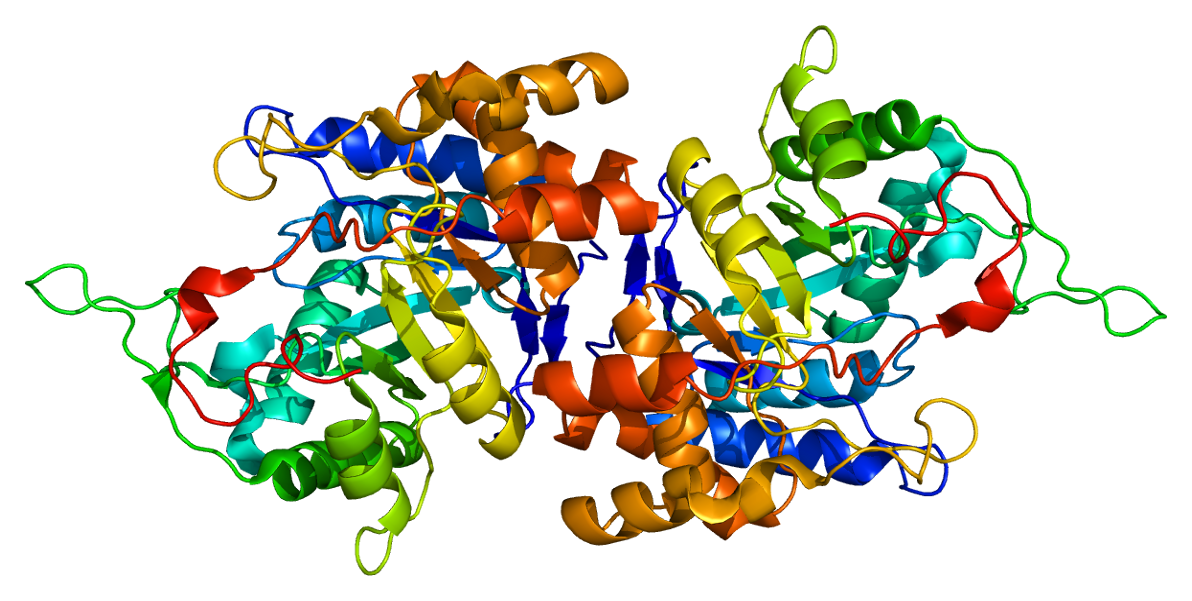
Aldo-keto Reductase
The aldo-keto reductase family is a family of proteins that are subdivided into 16 categories; these include a number of related monomeric NADPH-dependent oxidoreductases, such as aldehyde reductase, aldose reductase, prostaglandin F synthase, xylose reductase, rho crystallin, and many others. Structure All possess a similar structure, with a beta-alpha-beta fold characteristic of nucleotide binding proteins. The fold comprises a parallel beta-8/alpha-8-barrel, which contains a novel NADP-binding motif. The binding site is located in a large, deep, elliptical pocket in the C-terminal end of the beta sheet, the substrate being bound in an extended conformation. The hydrophobic nature of the pocket favours aromatic and apolar substrates over highly polar ones. Binding of the NADPH coenzyme causes a massive conformational change, reorienting a loop, effectively locking the coenzyme in place. This binding is more similar to FAD- than to NAD(P)-binding oxidoreductases. Examples S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribbon Diagram
Ribbon diagrams, also known as Richardson diagrams, are three-dimensional space, 3D schematic representations of protein structure and are one of the most common methods of protein depiction used today. The ribbon depicts the general course and organization of the protein backbone in 3D and serves as a visual framework for hanging details of the entire atomic structure, such as the balls for the oxygen atoms attached to myoglobin's active site in the adjacent figure. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. Alpha helix, α-helices are shown as coiled ribbons or thick tubes, Beta sheet, β-sheets as arrows, and non-repetitive coils or loops as lines or thin tubes. The direction of the Peptide, polypeptide chain is shown locally by the arrows, and may be indicated overall by a colour ramp along the length of the ribbon. Ribbon diagrams are simple yet powerful, expressing the visual basics of a molecular structure (twist, fold an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all Life, life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common Nutrient, nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a pentose, five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domains
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer after ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Steroidogenic Enzyme
__NOTOC__ Steroidogenic enzymes are enzymes that are involved in steroidogenesis and steroid biosynthesis. They are responsible for the biosynthesis of the steroid hormones, including sex steroids (androgens, estrogens, and progestogens) and corticosteroids (glucocorticoids and mineralocorticoids), as well as neurosteroids, from cholesterol. Steroidogenic enzymes are most highly expressed in classical steroidogenic tissues, such as the testis, ovary, and adrenal cortex, but are also present in other tissues in the body. List of steroidogenic enzymes * Steroid desmolases ** Cholesterol side-chain cleavage enzyme (20,22-desmolase) – steroid synthesis ** 17,20-Lyase (17,20-desmolase) – androgen synthesis * Steroid hydroxylases ** 11β-Hydroxylase – corticosteroid synthesis ** 17α-Hydroxylase – androgen and glucocorticoid synthesis ** 18-Hydroxylase (aldosterone synthase) – mineralocorticoid synthesis ** 21-Hydroxylase – corticosteroid synthesis ** Cytochrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AKR1
Aldo-keto reductase family 1 (AKR1) is a family of aldo-keto reductase enzymes that is involved in steroid metabolism. It includes the AKR1C and AKR1D subgroups, which respectively consist of AKR1C1– AKR1C4 and AKR1D1. Together with short-chain dehydrogenase/reductases (SDRs), these enzymes catalyze oxidoreductions, act on the C3, C5, C11, C17 and C20 positions of steroids, and function as , , 5β-reductases, , , and , respectively. The AKR1C enzymes act as 3-, 17- and 20- ketosteroid reductases, while AKR1D1 acts as the sole 5β-reductase in humans. Members AKR1A1; AKR1B1; AKR1B10; AKR1C1; AKR1C2; AKR1C3; AKR1C4; AKR1D1; Others See also * Steroidogenic enzyme __NOTOC__ Steroidogenic enzymes are enzymes that are involved in steroidogenesis and steroid biosynthesis. They are responsible for the biosynthesis of the steroid hormones, including sex steroids (androgens, estrogens, and progestogens) and co ... References Enzymes {{enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potassium Channel
Potassium channels are the most widely distributed type of ion channel found in virtually all organisms. They form potassium-selective pores that span cell membranes. Potassium channels are found in most cell types and control a wide variety of cell functions. Function Potassium channels function to conduct potassium ions down their electrochemical gradient, doing so both rapidly (up to the diffusion rate of K+ ions in bulk water) and selectively (excluding, most notably, sodium despite the sub-angstrom difference in ionic radius). Biologically, these channels act to set or reset the resting potential in many cells. In excitable cells, such as neurons, the delayed counterflow of potassium ions shapes the action potential. By contributing to the regulation of the cardiac action potential duration in cardiac muscle, malfunction of potassium channels may cause life-threatening arrhythmias. Potassium channels may also be involved in maintaining vascular tone. They also regu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ... or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
L-xylose 1-dehydrogenase
In enzymology, a L-xylose 1-dehydrogenase () is an enzyme that catalyzes the chemical reaction :L-xylose + NADP+ \rightleftharpoons L-xylono-1,4-lactone + NADPH + H+ Thus, the two substrates of this enzyme are L-xylose and NADP+, whereas its 3 products are L-xylono-1,4-lactone, NADPH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name A systematic name is a name given in a systematic way to one unique group, organism, object or chemical substance, out of a specific population or collection. Systematic names are usually part of a nomenclature. A semisystematic name or semitrivi ... of this enzyme class is L-xylose:NADP+ 1-oxidoreductase. Other names in common use include L-xylose dehydrogenase, and NADPH-xylose reductase. References * EC 1.1.1 NADPH-dependent enzymes Enzymes of unknown structure {{1.1.1-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nicotinamide Adenine Dinucleotide Phosphate
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP or, in older notation, TPN (triphosphopyridine nucleotide), is a Cofactor (biochemistry), cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the redox, reduced form, whereas NADP is the redox, oxidized form. NADP is used by all forms of cellular life. NADP is essential for life because it is needed for cellular respiration. NADP differs from NAD+, NAD by the presence of an additional phosphate group on the 2' position of the ribose ring that carries the adenine Moiety (chemistry), moiety. This extra phosphate is added by NAD+ kinase, NAD+ kinase and removed by NADP+ phosphatase. Biosynthesis NADP In general, NADP+ is synthesized before NADPH is. Such a reaction usually starts with NAD+, NAD+ from either the de-novo or the salvage pathway, with NAD+ kinase, NAD+ kinase adding the extra phosphate g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prostaglandin-F Synthase
In enzymology, a prostaglandin-F synthase (PGFS; ) is an enzyme that catalyzes the chemical reaction: :(5''Z'',13''E'')-(15''S'')-9alpha,11alpha,15-trihydroxyprosta-5,13-dienoate + NADP+ \rightleftharpoons (5''Z'',13''E'')-(15''S'')-9alpha,15-dihydroxy-11-oxoprosta-5,13-dienoate + NADPH + H+ Thus, the two products of this enzyme are 9α,11β–PGF2 and NADP+, whereas its three substrates are Prostaglandin D2, NADPH, and H+. PGFS is a monomeric wild-type protein that was first purified from bovine lung (PDB ID: 2F38). This enzyme belongs to the family of aldo-keto reductase (AKR) based on its high substrate specificity, its high molecular weight (38055.48 Da) and amino acid sequence. In addition, it is categorized as C3 (AKR1C3) because it is an isoform of 3α-hydroxysteroid dehydrogenase. The function of PGFS is to catalyze the reduction of aldehydes and ketones to their corresponding alcohols. In humans, these reactions take place mostly in the lungs and in the liver. Mor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
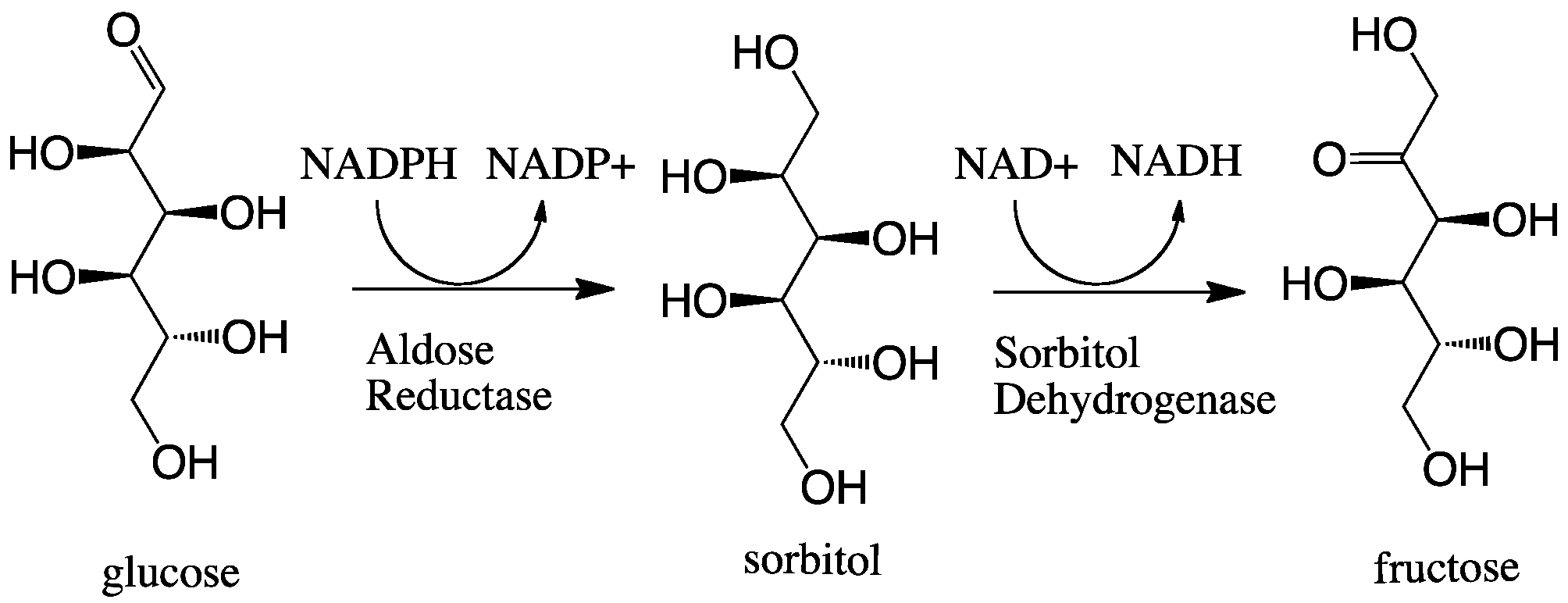
Aldose Reductase
In enzymology, aldose reductase (or aldehyde reductase) () is an enzyme in humans encoded by the gene AKR1B1. It is an cytosolic NADPH-dependent oxidoreductase that catalyzes the reduction of a variety of aldehydes and carbonyls, including monosaccharides, and primarily known for catalyzing the reduction of glucose to sorbitol, the first step in polyol pathway of glucose metabolism. Reactions Aldose reductase catalyzes the NADPH-dependent conversion of glucose to sorbitol, the first step in polyol pathway of glucose metabolism. The second and last step in the pathway is catalyzed by sorbitol dehydrogenase, which catalyzes the NAD-linked oxidation of sorbitol to fructose. Thus, the polyol pathway results in conversion of glucose to fructose with stoichiometric utilization of NADPH and production of NADH. ;glucose + NADPH + H+ \rightleftharpoons sorbitol + NADP+ Galactose is also a substrate for the polyol pathway, but the corresponding keto sugar is not produced because sorbito ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |