|
Proto-mitochondrion
The proto-mitochondrion is the hypothetical ancestral bacterial endosymbiont from which all mitochondria in eukaryotes are thought to descend, after an episode of symbiogenesis which created the aerobic eukaryotes. Phylogeny The phylogenetic analyses of the few genes that are still encoded in the genomes of modern mitochondria suggest an alphaproteobacterial origin for this endosymbiont, in an ancient episode of symbiogenesis early in the history of the eukaryotes. Although the order ''Rickettsiales'' has been proposed as the alphaproteobacterial sister-group of mitochondria, no definitive evidence pinpoints the alphaproteobacterial group from which the proto-mitochondrion emerged, and some contradictory evidence, especially in the early, sparse genome samplings. Martijn ''et al'' found mitochondria are a possible sister-group to all other alphaproteobacteria. The phylogenetic tree of the Rickettsidae has been inferred by Ferla ''et al.'' from the comparison of 16S + 23S riboso ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endosymbiont
An endosymbiont or endobiont is an organism that lives within the body or cells of another organism. Typically the two organisms are in a mutualism (biology), mutualistic relationship. Examples are nitrogen-fixing bacteria (called rhizobia), which live in the root nodules of legumes, single-cell algae inside Coral reef, reef-building corals, and bacterial endosymbionts that provide essential nutrients to insects. Endosymbiosis played key roles in the development of eukaryotes and plants. Roughly 2.2 billion years ago an archaeon absorbed a bacterium through phagocytosis, that eventually became the mitochondria that provide energy to almost all living Eukaryote, eukaryotic cells. Approximately 1 billion years ago, some of those cells absorbed cyanobacteria that eventually became chloroplasts, organelles that produce energy from sunlight. Approximately 100 million years ago, a lineage of amoeba in the genus ''Paulinella'' independently engulfed a cyanobacterium that evolved to be f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neorickettsia
''Neorickettsia'' is a genus of bacteria. Species or strains in this genus are coccoid or pleomorphic cells that reside in cytoplasmic vacuoles within monocytes and macrophages of dogs, horses, bats, and humans. '' Neorickettsia sennetsu'' causes Sennetsu ehrlichiosis. Unlike other forms of ehrlichiosis, ''Neorickettsia sennetsu'' is transmitted by trematode Trematoda is a Class (biology), class of flatworms known as trematodes, and commonly as flukes. They are obligate parasite, obligate Endoparasites, internal parasites with a complex biological life cycle, life cycle requiring at least two Host ( ...s from fish. '' Neorickettsia risticii'' causes Potomac horse fever. '' Neorickettsia helminthoeca'' is found in association with the trematode '' Nanophyetus salmincola'', and causes salmon poisoning disease in dogs and other canids. '' Neorickettsia elokominica'' causes a similar disease, Elokomin fluke fever, in canids and other species. Species * '' Neoricketts ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteome
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome. Types of proteomes While proteome generally refers to the proteome of an organism, multicellular organisms may have very different proteomes in different cells, hence it is important to distinguish proteomes in cells and organisms. A cellular proteome is the collection of proteins found in a particular cell (biology), cell type under a particular set of environmental conditions such as exposure to hormone, hormone stimulation. It can also be useful to consider an organism's complete proteome, which can be conceptualized as the complete set of proteins from all of the various cellular proteomes. This is very roughly the protein equivalent of the genome. The term ''proteome'' has also been used to r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coenzyme Q – Cytochrome C Reductase
The coenzyme Q : cytochrome ''c'' – oxidoreductase, sometimes called the cytochrome ''bc''1 complex, and at other times complex III, is the third complex in the electron transport chain (), playing a critical role in biochemical generation of ATP (oxidative phosphorylation). Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial (cytochrome b) and the nuclear genomes (all other subunits). Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most bacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits (cytochrome B, cytochrome C1, Rieske protein), 2 core proteins and 6 low-molecular weight proteins. Ubiquinol—cytochrome-c reductase catalyzes the chemical reaction :QH2 + 2 ferricytochrome c \rightleftharpoons Q + 2 ferrocytochrome c + 2 H+ Thus, the two substrates of this enzyme are qui ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but their discovery in eukaryotes was shown in the early 1990s, and are considered to be rare. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNAs. Operons are also found in viruses such as bacteriophages. For ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C Oxidase
The enzyme cytochrome c oxidase or Complex IV (was , now reclassified as a translocasEC 7.1.1.9 is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes. It is the last enzyme in the Cellular respiration, respiratory electron transport chain of cell (biology), cells located in the membrane. It receives an electron from each of four cytochrome c molecules and transfers them to one oxygen molecule and four protons, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton electrochemical potential, which the ATP synthase then uses to synthesize Adenosine triphosphate, ATP. Structure The complex The complex is a large integral membrane protein composed of several Cofactor (biochemistry)#Metal ions, metal prosthetic sites and 13 protein subunits in mammals. In mammals, ten subunits a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sphingolipid
Sphingolipids are a class of lipids containing a backbone of sphingoid bases, which are a set of aliphatic amino alcohols that includes sphingosine. They were discovered in brain extracts in the 1870s and were named after the mythological sphinx because of their enigmatic nature. These compounds play important roles in signal transduction and cell recognition. Sphingolipidoses, or disorders of sphingolipid metabolism, have particular impact on neural tissue. A sphingolipid with a terminal hydroxyl group is a ceramide. Other common groups bonded to the terminal oxygen atom include phosphocholine, yielding a sphingomyelin, and various sugar monomers or dimers, yielding cerebrosides and globosides, respectively. Cerebrosides and globosides are collectively known as glycosphingolipids. Structure The long-chain bases, sometimes simply known as sphingoid bases, are the first non-transient products of '' de novo'' sphingolipid synthesis in both yeast and mammals. These co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cardiolipin
Cardiolipin (IUPAC name 1,3-bis(''sn''-3’-phosphatidyl)-''sn''-glycerol, "''sn''" designating stereospecific numbering) is an important component of the inner mitochondrial membrane, where it constitutes about 20% of the total lipid composition. It can also be found in the membranes of most bacteria. The name "cardiolipin" is derived from the fact that it was first found in animal hearts. It was first isolated from the beef heart in the early 1940s by Mary C. Pangborn. In mammalian cells, but also in plant cells, cardiolipin (CL) is found almost exclusively in the inner mitochondrial membrane, where it is essential for the optimal function of numerous enzymes that are involved in mitochondrial energy metabolism. Structure Cardiolipin (CL) is a kind of diphosphatidylglycerol lipid. Two phosphatidic acid moieties connect with a glycerol backbone in the center to form a dimeric structure. So it has four alkyl groups and potentially carries two negative charges. As there are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
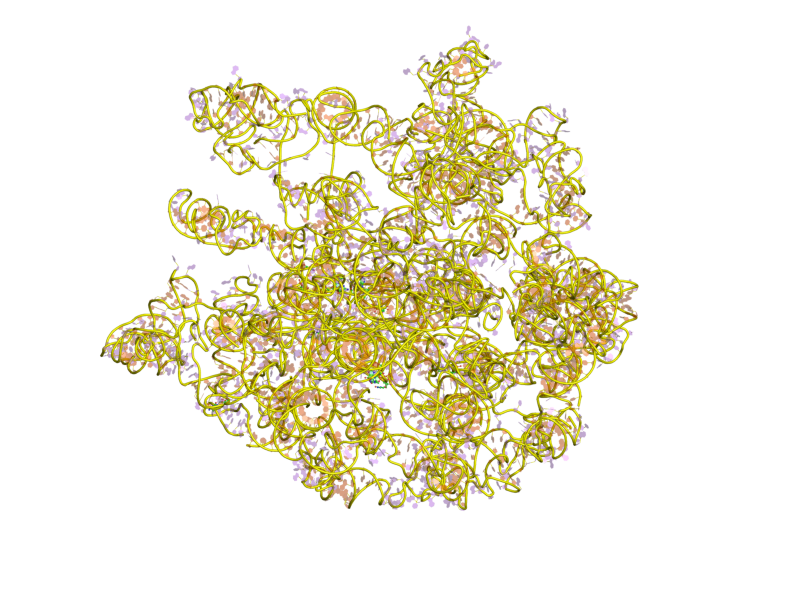
23S Ribosomal RNA
The 23S rRNA is a 2,904 nucleotide long (in ''E. coli'') component of the large subunit (50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains titled I-VI, with the corresponding 5S rRNA being considered domain VII. The ribosomal peptidyl transferase activity resides in domain V of this rRNA, which is also the most common binding site for antibiotics that inhibit translation, making it a target for ribosomal engineering. A well-known member of this antibiotic class, chloramphenicol, acts by inhibiting peptide bond formation, with recent 3D-structural studies showing two different binding sites depending on the species of ribosome. Numerous mutations in domains of the 23S rRNA with Peptidyl transferase activity have resulted in antibiotic resistance. 23S rRNA genes typically have higher sequence variations, including insertions and/or deletions, compared to other rRNAs. The eukary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA genes and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Terminology The descriptor ''16S'' refers to the size of these ribosomal subunits as reflected indirectly by the speed at which they sediment when samples are centrifuged. Thus ''16S'' means 16 Svedburg units. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogenophilalia
The ''Hydrogenophilaceae'' are a family of the class '' Hydrogenophilalia'' in the phylum ''Pseudomonadota'' ("Proteobacteria"), with two genera – '' Hydrogenophilus'' and '' Tepidiphilus''. Like all ''Pseudomonadota'', they are Gram-negative. All known species are thermophilic, growing around 50 °C, and use molecular hydrogen or organic molecules as their source of electrons to support growth; some species are autotrophs. The genus ''Thiobacillus'' was previously considered to be a member in this family but was reclassified into the order ''Nitrosomonadales'' at the same time that the '' Hydrogenophilales'' were removed from the ''Betaproteobacteria'' to form the class '' Hydrogenophilalia''. '' Hydrogenophilus thermoluteolus'' is a facultative chemolithoautotroph originally isolated from a hot spring; however, it was detected 2004 in ice core samples retrieved from a depth around 3 km within the ice covering Lake Vostok in Antarctica. The presence of DNA from (and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Betaproteobacteria
''Betaproteobacteria'' are a class of Gram-negative bacteria, and one of the six classes of the phylum '' Pseudomonadota'' (synonym Proteobacteria). Metabolism The ''Betaproteobacteria'' comprise over 75 genera and 400 species. Together, they represent a broad variety of metabolic strategies and occupy diverse environments, ranging from obligate pathogens living within host organisms to oligotrophic groundwater ecosystems. Whilst most members of the ''Betaproteobacteria'' are heterotrophic, deriving both their carbon and electrons from organocarbon sources, some are photoheterotrophic, deriving energy from light and carbon from organocarbon sources. Other genera are autotrophic, deriving their carbon from bicarbonate or carbon dioxide and their electrons from reduced inorganic ions such as nitrite, ammonium, thiosulfate or sulfide — many of these chemolithoautotrophic. ''Betaproteobacteria'' are economically important, with roles in maintaining soil pH and in elementa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |