|
SWISS-MODEL
Swiss-model (stylized as SWISS-MODEL) is a structural bioinformatics web-server dedicated to homology modeling of 3D protein structures. , homology modeling is the most accurate method to generate reliable three-dimensional protein structure models and is routinely used in many practical applications. Homology (or comparative) modelling methods make use of experimental protein structures (templates) to build models for evolutionary related proteins (targets). Today, Swiss-model consists of three tightly integrated components: (1) The Swiss-model pipeline – a suite of software tools and databases for automated protein structure modelling, (2) The Swiss-model Workspace – a web-based graphical user interface workbench, (3) The Swiss-model Repository – a continuously updated database of homology models for a set of model organism proteomes of high biomedical interest. Pipeline Swiss-model pipeline comprises the four main steps that are involved in building a homology model of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology Modelling
Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the "''target''" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the "''template''"). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of a sequence alignment that maps residues in the query sequence to residues in the template sequence. It has been seen that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure. Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure. It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Bioinformatics
Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term ''structural'' has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge. Introduction Protein structure The structure of a protein is directly related to its function. The presence of certa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caenorhabditis Elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''rhabditis'' (rod-like) and Latin ''elegans'' (elegant). In 1900, Émile Maupas, Maupas initially named it ''Rhabditidae, Rhabditides elegans.'' Günther Osche, Osche placed it in the subgenus ''Caenorhabditis'' in 1952, and in 1955, Ellsworth Dougherty, Dougherty raised ''Caenorhabditis'' to the status of genus. ''C. elegans'' is an unsegmented pseudocoelomate and lacks respiratory or circulatory systems. Most of these nematodes are hermaphrodites and a few are males. Males have specialised tails for mating that include spicule (nematode), spicules. In 1963, Sydney Brenner proposed research into ''C. elegans,'' primarily in the area of neuronal development. In 1974, he began research into the molecular biology, molecular and developmental ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CASP
Critical Assessment of Structure Prediction (CASP), sometimes called Critical Assessment of Protein Structure Prediction, is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994. CASP provides research groups with an opportunity to objectively test their structure prediction methods and delivers an independent assessment of the state of the art in protein structure modeling to the research community and software users. Even though the primary goal of CASP is to help advance the methods of identifying protein three-dimensional structure from its amino acid sequence many view the experiment more as a "world championship" in this field of science. More than 100 research groups from all over the world participate in CASP on a regular basis and it is not uncommon for entire groups to suspend their other research for months while they focus on getting their servers ready for the experiment and on performing the detailed predictio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Prediction Software
This list of protein structure prediction software summarizes notable used software tools in protein structure prediction, including homology modeling, protein threading, ''ab initio'' methods, Protein structure prediction#Secondary structure, secondary structure prediction, and transmembrane helix and signal peptide prediction. Software list Below is a list which separates programs according to the method used for structure prediction. Homology modeling Threading and fold recognition ''Ab initio'' structure prediction Secondary structure prediction Detailed list of programs can be found at List of protein secondary structure prediction programs See also *List of protein secondary structure prediction programs *Comparison of nucleic acid simulation software *List of software for molecular mechanics modeling *Molecular design software *Protein design *AlphaFold External linksbio.tools finding more tools References {{DEFAULTSORT:Protein structure predictio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure from Protein primary structure, primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology and addresses Levinthal's paradox. Accurate structure prediction has important applications in medicine (for example, in drug design) and biotechnology (for example, in novel enzyme design). Starting in 1994, the performance of current methods is assessed biannually in the ''Critical Assessment of Structure Prediction'' (CASP) experiment. A continuous evaluation of protein structure prediction web servers is performed by the community project ''Continuous Automated Model EvaluatiOn'' (CAMEO3D). Protein structure a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CAMEO3D
Continuous Automated Model EvaluatiOn (CAMEO) is a community-wide project to continuously evaluate the accuracy and reliability of protein structure prediction servers in a fully automated manner. CAMEO is a continuous and fully automated complement to the bi-annual CASP experiment. Currently, CAMEO evaluates predictions for predicted three-dimensional protein structures (3D), ligand binding site predictions in proteins (LB), and model quality estimation tools (QE). Workflow CAMEO performs blind assessment of protein structure prediction techniques based on the weekly releases of newly determined experimental structures by the Protein Databank (PDB). The amino acid sequences of soon to be released protein structures are submitted to the participating web-servers. The web-servers return their predictions to CAMEO, and predictions received before the experimental structures have been released are included in the assessment of prediction accuracy. In contrast to the CASP experimen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EVA (benchmark)
EVA was a continuously running benchmark project for assessing the quality and value of protein structure prediction and secondary structure prediction methods. Methods for predicting both secondary structure and tertiary structure - including homology modeling, protein threading, and contact order prediction - were compared to results from each week's newly solved protein structures deposited in the Protein Data Bank. The project aimed to determine the prediction accuracy that would be expected for non-expert users of common, publicly available prediction webservers; this is similar to the related LiveBench project and stands in contrast to the bi-yearly benchmark CASP Critical Assessment of Structure Prediction (CASP), sometimes called Critical Assessment of Protein Structure Prediction, is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994. CASP pro ..., which aims to identify the maximum accuracy achievable by pred ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
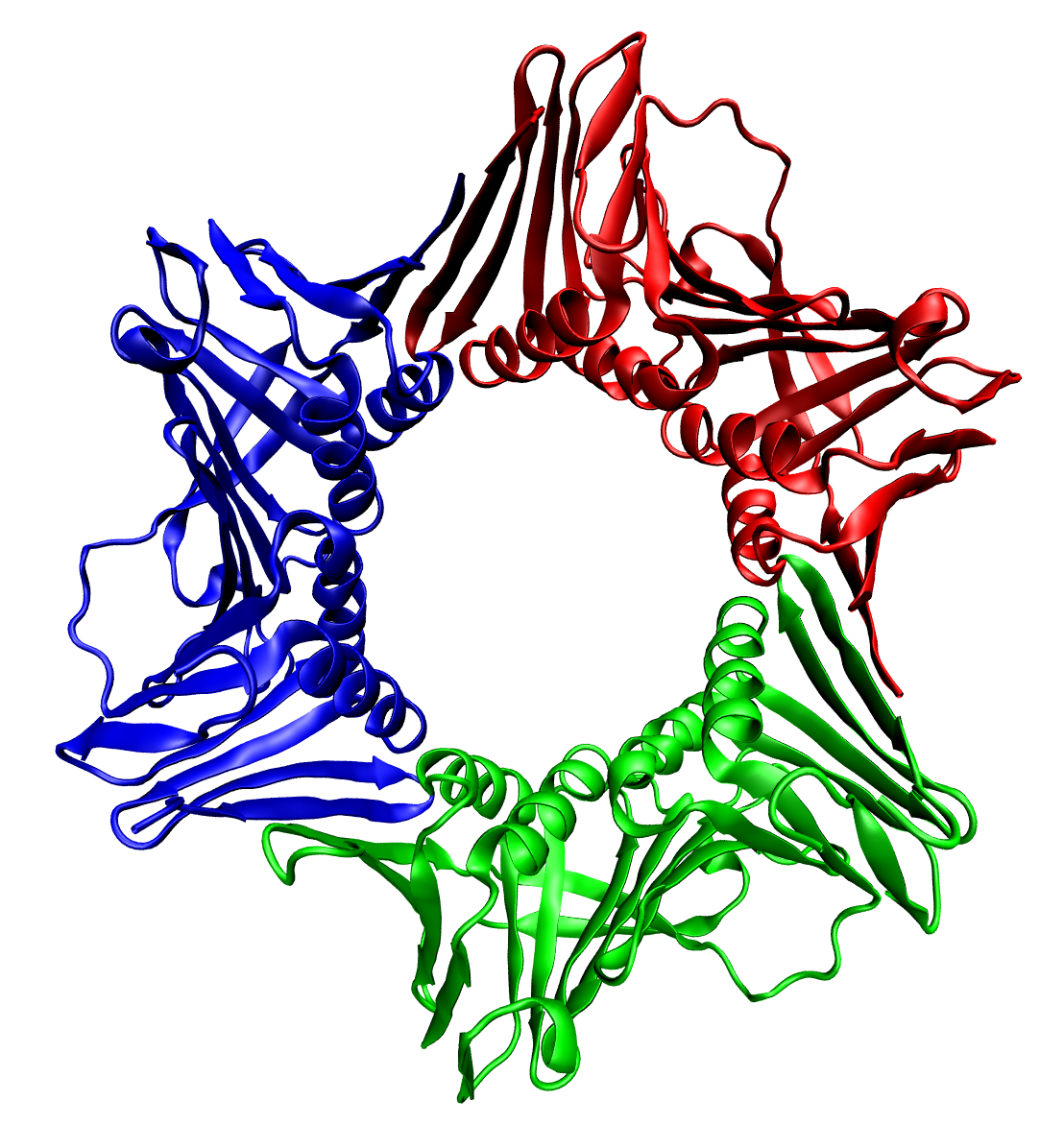
Protein Quaternary Structure
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors. Description and examples Many proteins are actually assemblies of multiple polypeptide chains. The quaternary structure refers to the number and arrangement of the protein subunits w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Initiative
The Protein Structure Initiative (PSI) was a USA based project that aimed at accelerating discovery in structural genomics and contribute to understanding biological function. Funded by the U.S. National Institute of General Medical Sciences (NIGMS) between 2000 and 2015, its aim was to reduce the cost and time required to determine three-dimensional protein structures and to develop techniques for solving challenging problems in structural biology, including membrane proteins. Over a dozen research centers have been supported by the PSI for work in building and maintaining high-throughput structural genomics pipelines, developing computational protein structure prediction methods, organizing and disseminating information generated by the PSI, and applying high-throughput structure determination to study a broad range of important biological and biomedical problems. The project has been organized into three separate phases. The first phase of the Protein Structure Initiative (PSI- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STRING
String or strings may refer to: *String (structure), a long flexible structure made from threads twisted together, which is used to tie, bind, or hang other objects Arts, entertainment, and media Films * ''Strings'' (1991 film), a Canadian animated short * ''Strings'' (2004 film), a film directed by Anders Rønnow Klarlund * ''Strings'' (2011 film), an American dramatic thriller film * ''Strings'' (2012 film), a British film by Rob Savage * '' Bravetown'' (2015 film), an American drama film originally titled ''Strings'' * '' The String'' (2009), a French film Music Instruments * String (music), the flexible element that produces vibrations and sound in string instruments * String instrument, a musical instrument that produces sound through vibrating strings ** List of string instruments * String piano, a pianistic extended technique in which sound is produced by direct manipulation of the strings, rather than striking the piano's keys Types of groups * String band, music ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |