|
Small Nucleolar RNA R66
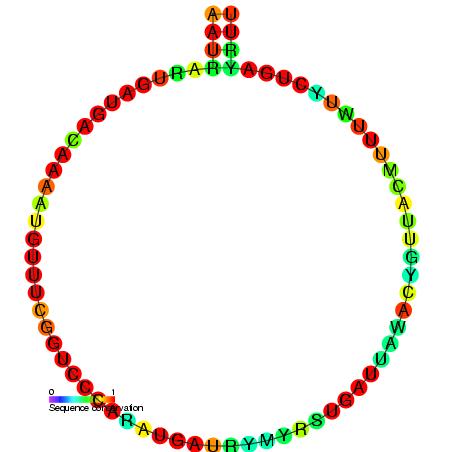
In molecular biology, Small nucleolar RNA R66 (also known as snoR66) is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoR66 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoR66 was identified by a computational screening of the rice ''Oryza sativa'' genome and is proposed to acts as a methylation guide for 18S ribosomal RNA in plants. Rice snoR66 has also been alternatively named snoZ269. Its should also not be confused with the snoRNA identified in yeast (''Saccharomyces cerevisiae ''Saccharomyces ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnoRNA
In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing. snoRNA guided modifications After transcription, nascent rRNA molecules (termed pre-rRNA) undergo a series of processing steps to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases, the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs. *Methylation is the attachment or substitution of a methyl group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Nucleolar RNA SnR66
snoRNA snR66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR66 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ... of substrate RNAs. snoRNA snR66 was initially discovered using a computational screen of the ''Saccharomyces cerevisiae'' genome. References External li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like '' Escherichia coli'' as the model bacterium. It is the microorganism which causes many common types of fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley bodies present, which are involved in particular secretory pathways. Antibodies again ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oryza Sativa
''Oryza sativa'', having the common name Asian cultivated rice, is the much more common of the two rice species cultivated as a cereal, the other species being ''Oryza glaberrima, O. glaberrima'', African rice. It was History of rice cultivation, first domesticated in the Yangtze River basin in China 13,500 to 8,200 years ago. ''Oryza sativa'' belongs to the genus ''Oryza'' and the BOP clade in the grass family Poaceae. With a genome consisting of 430megabase, Mbp across 12 chromosomes, it is renowned for being easy to Genetically modified rice, genetically modify and is a model organism for the study of the biology of cereals and Monocotyledon, monocots. Description ''O. sativa'' has an erect stalk stem that grows tall, with a smooth surface. The leaf is lanceolate, long, and grows from a ligule long. Image:Kerbau Jawa.jpg, Domestic buffalo, Water buffalo ploughing a rice paddyfield, Java File:Jumli Marshi Oryza sativa Rice.jpg, Jumli Marshi, brown rice from Nepal File: ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular And Cellular Biology
''Molecular and Cellular Biology'' is a biweekly peer-reviewed scientific journal covering all aspects of molecular and cellular biology. It is published by the American Society for Microbiology and the editor-in-chief is Peter Tontonoz (University of California, Los Angeles). It was established in 1981. The h-index (1981-2021) is 338. Abstracting and indexing The journal is abstracted and indexed in: According to the ''Journal Citation Reports'', the journal has a 2021 impact factor The impact factor (IF) or journal impact factor (JIF) of an academic journal is a type of journal ranking. Journals with higher impact factor values are considered more prestigious or important within their field. The Impact Factor of a journa ... of 5.069. References External links * Molecular and cellular biology journals Delayed open access journals Academic journals established in 1981 English-language journals Biweekly journals American Society for Microbiology academic jour ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen#Compounds, hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and biology. In biological systems, methylation is Catalysis, catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of Protein#Functions, protein function, and RNA processing. ''In vitro'' methylation of tissue samples is also a way to reduce some histology#Histological Artifacts, histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals and can regulate gene expression, RNA processing, and protein function. It is a key pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnoRNA
In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing. snoRNA guided modifications After transcription, nascent rRNA molecules (termed pre-rRNA) undergo a series of processing steps to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases, the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs. *Methylation is the attachment or substitution of a methyl group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolus
The nucleolus (; : nucleoli ) is the largest structure in the cell nucleus, nucleus of eukaryote, eukaryotic cell (biology), cells. It is best known as the site of ribosome biogenesis. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of the nucleolus is the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Theodor Schwann in his 1839 treatise described that Matthias Jakob Schleiden, Schleiden had identified small corpuscles in nuclei, and named the structures "Kernkörperchen". In a 1947 translation of the work to English, the structure was named "nucleolus". Little was known about the fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids (DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst eukaryotes, and the tmRNA in bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Nuclear RNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the Cell nucleus#Splicing speckles, splicing speckles and Cajal body, Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors (7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres. snRNA are always associated with a set of specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP, often pronounced "snurps"). Each snRNP particle is composed of a snRNA component and several snRNP-specific proteins (including LSm, Sm proteins, a family of nuclear proteins). The most common human snRNA components of these complexes are known, respectively, as: U1 spliceos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important list of RNAs, types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, Extracellular RNA, exRNAs, scaRNAs and the long noncoding RNA, long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent Transcriptomics, transcriptomic and Bioinformatics, bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have unknown functions, if any. There is no consensus on how much of non-coding transcription is functional: some believe most ncRNAs to be non-functional "junk RNA", spurious transcriptions, while others expect that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |