|
Restriction Site Associated DNA Markers
Restriction site associated DNA (RAD) markers are a type of genetic marker which are useful for association mapping, QTL-mapping, population genetics, ecological genetics and evolutionary genetics. The use of RAD markers for genetic mapping is often called RAD mapping. An important aspect of RAD markers and mapping is the process of isolating RAD tags, which are the DNA sequences that immediately flank each instance of a particular restriction site of a restriction enzyme throughout the genome. Once RAD tags have been isolated, they can be used to identify and genotype DNA sequence polymorphisms mainly in form of single nucleotide polymorphisms (SNPs). Polymorphisms that are identified and genotyped by isolating and analyzing RAD tags are referred to as RAD markers. Although genotyping by sequencing presents an approach similar to the RAD-seq method, they differ in some substantial ways. Isolation of RAD tags The use of the flanking DNA sequences around each restriction site is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Blunt Ends
DNA ends refer to the properties of the ends of linear DNA molecules, which in molecular biology are described as "sticky" or "blunt" based on the shape of the complementary strands at the terminus. In sticky ends, one strand is longer than the other (typically by at least a few nucleotides), such that the longer strand has bases which are left unpaired. In blunt ends, both strands are of equal length – i.e. they end at the same base position, leaving no unpaired bases on either strand. The concept is used in molecular biology, in cloning, or when subcloning insert DNA into vector DNA. Such ends may be generated by restriction enzymes that break the molecule's phosphodiester backbone at specific locations, which themselves belong to a larger class of enzymes called exonucleases and endonucleases. A restriction enzyme that cuts the backbones of both strands at non-adjacent locations leaves a staggered cut, generating two overlapping sticky ends, while an enzyme that makes a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arthropod
Arthropods ( ) are invertebrates in the phylum Arthropoda. They possess an arthropod exoskeleton, exoskeleton with a cuticle made of chitin, often Mineralization (biology), mineralised with calcium carbonate, a body with differentiated (Metamerism (biology), metameric) Segmentation (biology), segments, and paired jointed appendages. In order to keep growing, they must go through stages of moulting, a process by which they shed their exoskeleton to reveal a new one. They form an extremely diverse group of up to ten million species. Haemolymph is the analogue of blood for most arthropods. An arthropod has an open circulatory system, with a body cavity called a haemocoel through which haemolymph circulates to the interior Organ (anatomy), organs. Like their exteriors, the internal organs of arthropods are generally built of repeated segments. They have ladder-like nervous systems, with paired Anatomical terms of location#Dorsal and ventral, ventral Ventral nerve cord, nerve cord ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regent Honeyeater
The regent honeyeater (''Anthochaera phrygia'') is a critically endangered bird endemic to southeastern Australia. It is commonly considered a flagship species within its range, with the efforts going into its conservation having positive effects on many other species that share its habitat. Recent genetic research suggests it is closely related to the wattlebirds. Taxonomy First described by the English naturalist George Shaw (biologist), George Shaw in 1794, the regent honeyeater was moved to ''Anthochaera'' in 1827 by the naturalists Nicholas Aylward Vigors and Thomas Horsfield. It was known as ''Xanthomyza phrygia'' (Zanthomiza by Gregory Mathews)for many years, the genus erected by William Swainson in 1837. DNA analysis shows that its ancestry is in fact nested within the wattlebird genus ''Anthochaera''. The ancestor of the regent honeyeater split from a lineage that gave rise to the red wattlebird, red and yellow wattlebirds. The little and western wattlebirds arose from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
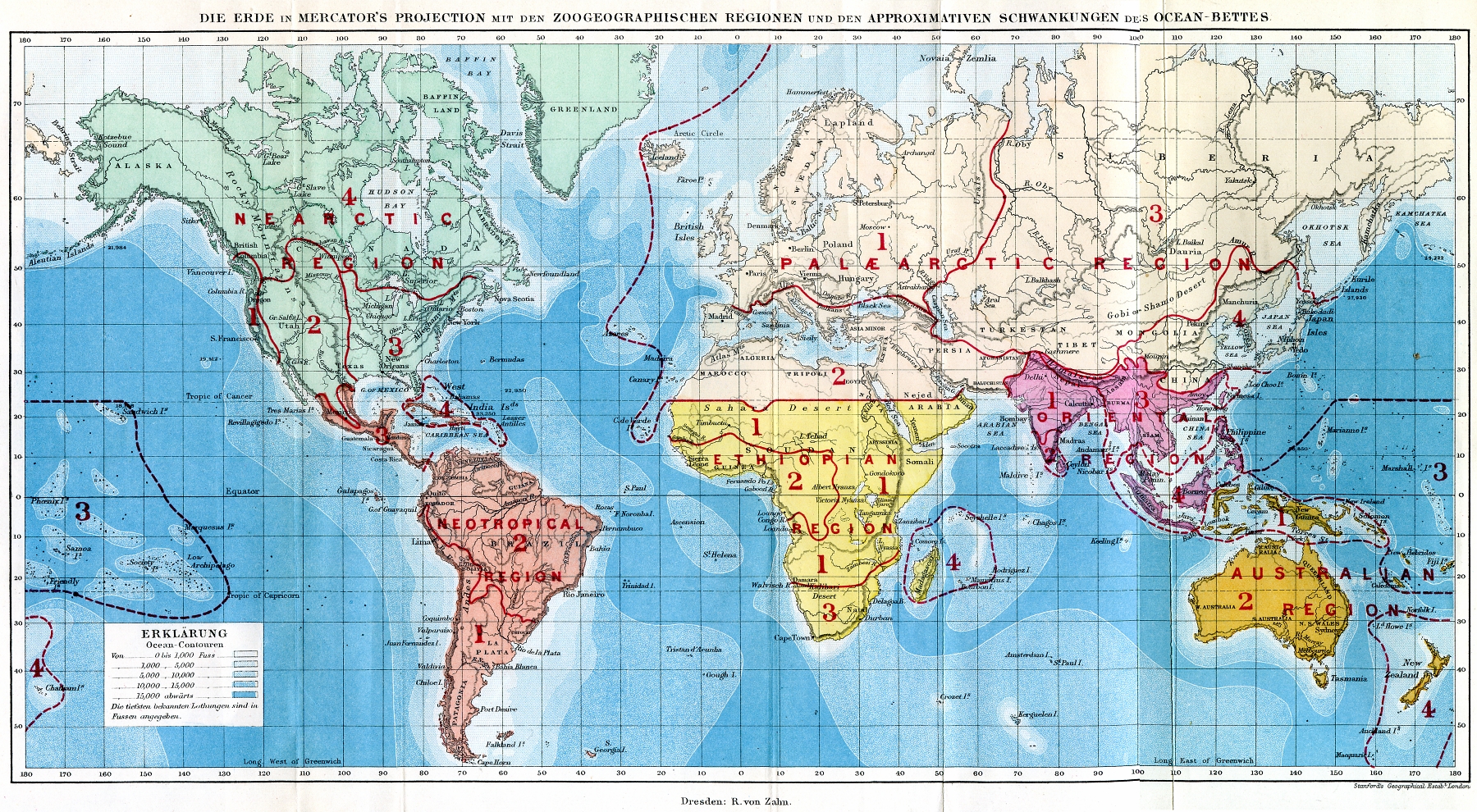
Palearctic Realm
The Palearctic or Palaearctic is a biogeographic realm of the Earth, the largest of eight. Confined almost entirely to the Eastern Hemisphere, it stretches across Europe and Asia, north of the foothills of the Himalayas, and North Africa. The realm consists of several bioregions: the Mediterranean Basin; North Africa; North Arabia; Western, Central and East Asia. The Palaearctic realm also has numerous rivers and lakes, forming several freshwater ecoregions. Both the eastern and westernmost extremes of the Paleartic span into the Western Hemisphere, including Cape Dezhnyov in Chukotka Autonomous Okrug to the east and Iceland to the west. The term was first used in the 19th century, and is still in use as the basis for zoogeographic classification. History In an 1858 paper for the ''Proceedings of the Linnean Society'', British zoologist Philip Sclater first identified six terrestrial zoogeographic realms of the world: Palaearctic, Aethiopian/ Afrotropic, Indian/ Indom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oedaleus Decorus
''Oedaleus'' is a genus of grasshoppers in the family Acrididae Acrididae are the predominant family of grasshoppers, comprising some 10,000 of the 11,000 species of the entire suborder Caelifera. The Acrididae are best known because all locusts (swarming grasshoppers) are of the Acrididae. The subfamily Oedi .... Species The ''Orthoptera Species File'' lists the following species: * '' Oedaleus abruptus'' (Thunberg, 1815) * '' Oedaleus australis'' Saussure, 1888 * '' Oedaleus bimaculatus'' Zheng & Gong, 2001 * '' Oedaleus carvalhoi'' Bolívar, 1889 * '' Oedaleus cnecosopodius'' Zheng, 2000 * '' Oedaleus decorus'' (Germar, 1825) * '' Oedaleus flavus'' (Linnaeus, 1758) * '' Oedaleus formosanus'' (Shiraki, 1910) * '' Oedaleus hyalinus'' Zheng & Mao, 1997 * '' Oedaleus immaculatus'' Chopard, 1957 * '' Oedaleus infernalis'' Saussure, 1884 * '' Oedaleus inornatus'' Schulthess Schindler, 1898 * '' Oedaleus instillatus'' Burr, 1900 * '' Oedaleus interruptus'' (Kirby, 1902) * '' Oedale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Site
In molecular biology, restriction sites, or restriction recognition sites, are regions of a DNA molecule containing specific (4-8 base pairs in length) sequences of nucleotides; these are recognized by restriction enzymes, which cleave the DNA at or near the site. These are generally palindromic sequences (because restriction enzymes usually bind as homodimers), and a particular restriction enzyme may cut the sequence between two nucleotides within its recognition site, or somewhere nearby. Function For example, the common restriction enzyme EcoRI recognizes the palindromic sequence GAATTC and cuts between the G and the A on both the top and bottom strands. This leaves an overhang (an end-portion of a DNA strand with no attached complement) known as a sticky end on each end of AATT. The overhang can then be used to ligate in (see DNA ligase) a piece of DNA with a complementary overhang (another EcoRI-cut piece, for example). Some restriction enzymes cut DNA at a restriction sit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Museomics
Museomics is the study of genomic data obtained from ancient DNA (aDNA) and historic DNA (hDNA) specimens in museum collections. Early research in this area focused on short sequences of DNA from mitochondrial genes, but sequencing of whole genomes has become possible. Next-generation sequencing (NGS) and high-throughput sequencing (HTS) methods can be applied to the analysis of genetic datasets extracted from collections materials. Such techniques have been described as a "third revolution in sequencing technology". Like radiocarbon dating, the techniques of museomics are a transformative technology. Results are revising and sometimes overturning previously accepted theories about a wide variety of topics such as the domestication of the horse. Museum collections contain unique resources such as natural history specimens, which can be used for genome-scale examinations of species, their evolution, and their responses to environmental change. Ancient DNA provides a unique windo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Homology
Sequence homology is the homology (biology), biological homology between DNA sequence, DNA, RNA sequence, RNA, or Protein primary structure, protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a Gene duplication, duplication event (paralogs), or else a Horizontal gene transfer, horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Sequence alignment, Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shotgun Sequencing
In genetics, shotgun sequencing is a method used for sequencing random DNA strands. It is named by analogy with the rapidly expanding, quasi-random shot grouping of a shotgun. The Sanger sequencing#Method, chain-termination method of DNA sequencing ("Sanger sequencing") can only be used for short DNA strands of 100 to 1000 base pairs. Due to this size limit, longer sequences are subdivided into smaller fragments that can be sequenced separately, and these sequences are sequence assembly, assembled to give the overall sequence. In shotgun sequencing, DNA is broken up randomly into numerous small segments, which are sequenced using the chain termination method to obtain ''reads''. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence. Shotgun sequencing was one of the precursor technologies that was resp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Population Differentiation
Population is a set of humans or other organisms in a given region or area. Governments conduct a census to quantify the resident population size within a given jurisdiction. The term is also applied to non-human animals, microorganisms, and plants, and has specific uses within such fields as ecology and genetics. Etymology The word ''population'' is derived from the Late Latin ''populatio'' (a people, a multitude), which itself is derived from the Latin word ''populus'' (a people). Use of the term Social sciences In sociology and population geography, population refers to a group of human beings with some predefined feature in common, such as location, race, ethnicity, nationality, or religion. Ecology In ecology, a population is a group of organisms of the same species which inhabit the same geographical area and are capable of interbreeding. The area of a sexual population is the area where interbreeding is possible between any opposite-sex pair within the area ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
University Of Oregon
The University of Oregon (UO, U of O or Oregon) is a Public university, public research university in Eugene, Oregon, United States. Founded in 1876, the university is organized into nine colleges and schools and offers 420 undergraduate and graduate degree programs. The university also operates the Ballmer Institute for Children's Behavioral Health in Portland, Oregon; the Oregon Institute of Marine Biology in Charleston, Oregon; and Pine Mountain Observatory in Central Oregon. UO's 295-acre campus is situated along the Willamette River. Most academic programs follow the 10-week quarter system. The university is Carnegie Classification of Institutions of Higher Education, classified among "R1: Doctoral Universities – Very high research activity" and is a member of the Association of American Universities. Since July 2014, UO has been governed by Board of Trustees of the University of Oregon, its own board of trustees. UO student-athletes compete as the Oregon Ducks and are pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |