|
RGG Box
The arginine-glycine or arginine-glycine-glycine (RG/RGG) motif is a repeating amino acid sequence motif commonly found in RNA-binding proteins (RBPs). RGG regions in proteins are defined as two or more RG/RGG sequences within a stretch of 30 amino acids. Initially named the RGG box, it confers a protein with the ability to bind double-stranded mRNA molecules. The RGG motif has been observed in proteins from at least 12 animal species, including humans. Biochemical function RGG motifs are primarily involved in mediating protein-RNA interactions. Positive charges from arginine residues promote electrostatic interactions with mRNA molecules. The composition and structure of the arginine side chain may also allow for specific interactions with other molecules as opposed to the other positively charged amino acids, lysine and histidine. Glycine residues add flexibility to the peptide structure and promote their tendency to form intrinsically disordered regions. The RGG motif can al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups ( alpha- , beta- , gamma- amino acids, etc.); other categories relate to polarity, ionization, and side-chain group type ( aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino-acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence. Amino acids are formally named by the IUPAC- IUBMB Joint Commi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
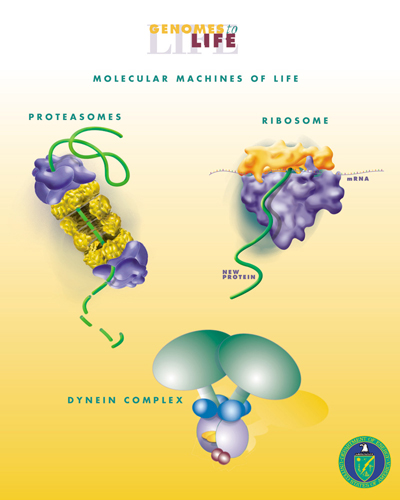
Cellular Engineering
Biological engineering or bioengineering is the application of principles of biology and the tools of engineering to create usable, tangible, economically viable products. Biological engineering employs knowledge and expertise from a number of pure and applied sciences, such as mass and heat transfer, kinetics, biocatalysts, biomechanics, bioinformatics, separation and purification processes, bioreactor design, surface science, fluid mechanics, thermodynamics, and polymer science. It is used in the design of medical devices, diagnostic equipment, biocompatible materials, renewable energy, ecological engineering, agricultural engineering, process engineering and catalysis, and other areas that improve the living standards of societies. Examples of bioengineering research include bacteria engineered to produce chemicals, new medical imaging technology, portable and rapid disease diagnostic devices, prosthetics, biopharmaceuticals, and tissue-engineered organs. Bioengine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FUS (gene)
RNA-binding protein fused in sarcoma/translocated in liposarcoma (FUS/TLS), also known as heterogeneous nuclear ribonucleoprotein P2 is a protein that in humans is encoded by the ''FUS'' gene. Discovery FUS/TLS was initially identified as a fusion protein (FUS-CHOP) produced as a result of chromosomal translocations in human cancers, especially liposarcomas. In these instances, the promoter and N-terminal part of FUS/TLS is translocated to the C-terminal domain of various DNA-binding transcription factors (e.g. C/EBP homologous protein, CHOP) conferring a strong transcriptional activation domain onto the fusion proteins. FUS/TLS was independently identified as the hnRNP P2 protein, a subunit of a complex involved in the maturation of pre-mRNA. Structure FUS/TLS is a member of the FET protein family that also includes the Ewing sarcoma breakpoint region 1, EWS protein, the TATA-binding protein TBP-associated factor TAFII68/TAF15, and the Drosophila cabeza/SARF protein. FUS/T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FMR1
''FMR1'' (Fragile X Messenger Ribonucleoprotein 1) is a human gene that codes for a protein called ''fragile X messenger ribonucleoprotein'', or FMRP. This protein, most commonly found in the brain, is essential for normal cognitive development and female reproductive function. Mutations of this gene can lead to fragile X syndrome, intellectual disability, premature ovarian failure, autism, Parkinson's disease, developmental delays and other cognitive deficits. The FMR1 premutation is associated with a wide spectrum of clinical phenotypes that affect more than two million people worldwide. Function Synaptic plasticity FMRP has a diverse array of functions throughout different areas of the neuron; however these functions have not been fully characterized. FMRP has been suggested to play roles in nucleocytoplasmic shuttling of mRNA, dendritic mRNA localization, and synaptic protein synthesis. Studies of Fragile X syndrome have significantly aided in the understanding of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolic Engineering
Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cell's production of a certain substance. These processes are chemical networks that use a series of biochemical reactions and enzymes that allow cells to convert raw materials into molecules necessary for the cell's survival. Metabolic engineering specifically seeks to mathematically model these networks, calculate a yield of useful products, and pin point parts of the network that constrain the production of these products. Genetic engineering techniques can then be used to modify the network in order to relieve these constraints. Once again this modified network can be modeled to calculate the new product yield. The ultimate goal of metabolic engineering is to be able to use these organisms to produce valuable substances on an industrial scale in a cost-effective manner. Current examples include producing beer, wine, cheese, pharmaceuticals, and other biotechnolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phase Separation
Phase separation is the creation of two distinct Phase (matter), phases from a single homogeneous mixture. The most common type of phase separation is between two immiscible liquids, such as oil and water. This type of phase separation is known as liquid-liquid equilibrium. Colloids are formed by phase separation, though not all phase separations forms colloids - for example oil and water can form separated layers under gravity rather than remaining as microscopic droplets in suspension. A common form of spontaneous phase separation is termed spinodal decomposition; it is described by the Cahn–Hilliard equation. Regions of a phase diagram in which phase separation occurs are called miscibility gaps. There are two boundary curves of note: the binodal, binodal coexistence curve and the spinodal, spinodal curve. On one side of the binodal, mixtures are absolutely stable. In between the binodal and the spinodal, mixtures may be metastable: staying mixed (or unmixed) absent some ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the " structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for example ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered protein tertiary structure, three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like Protein aggregation, aggregates, or flexible linkers in large multi-Protein domain, domain proteins. They are sometimes considered as a separate class of proteins along with globular protein, globular, fibrous protein, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins. They are most numerous in Eukaryote, eukaryotes, with an estimated 30-40% of residues in the eukaryotic proteome located in disordered regions. Disorder is present in around 70% of proteins, either in the form of disordered tails or flexible linkers. Proteins can also be entirely disordered and lack a define ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-covalent Interaction
In chemistry, a non-covalent interaction differs from a covalent bond in that it does not involve the sharing of electrons, but rather involves more dispersed variations of electromagnetic interactions between molecules or within a molecule. The chemical energy released in the formation of non-covalent interactions is typically on the order of 1–5 kcal/ mol (1000–5000 calories per 6.02 molecules). Non-covalent interactions can be classified into different categories, such as electrostatic, π-effects, van der Waals forces, and hydrophobic effects. Non-covalent interactions are critical in maintaining the three-dimensional structure of large molecules, such as proteins and nucleic acids. They are also involved in many biological processes in which large molecules bind specifically but transiently to one another (see the properties section of the DNA page). These interactions also heavily influence drug design, crystallinity and design of materials, particularly for self-assemb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |