|
P Element
''P'' elements are transposable elements that were discovered in ''Drosophila'' as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the ''P'' trait of the ''P'' element and it is found only in wild flies. They are also found in many other eukaryotes. The name was first suggested by evolutionary biologist Margaret Kidwell, who, together with James Kidwell and John Sved, researched hybrid dysgenesis in ''Drosophila''. They referred to strains as P of paternal and M of maternal if they contributed to hybrid dysgenesis in this reproductive role. The ''P'' element encodes for an enzyme known as ''P'' transposase. Unlike laboratory-bred females, wild-type females are thought also to express an inhibitor to ''P'' transposase function, produced by the very same element. This inhibitor reduces the disruption to the genome caused by the movement of ''P'' elements, allowing fertile progeny. Evidence for this comes from crosses of laboratory ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposable Elements
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome. The discovery of mobile genetic elements earned Barbara McClintock a Nobel Prize in 1983. There are at least two classes of TEs: Class I TEs or retrotransposons generally function via reverse transcription, while Class II TEs or DNA transposons encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. Discovery by Barbara McClintock Barbara McClintock discovered the first TEs in maize (''Zea mays'') at the Cold Spring Harbor Laboratory in New York. McClintock was experimenting with maize plants that had broken chromosomes. In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (style) of the flower received pollen from its own anther. These kernels came from a long line ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Splicing (genetics)
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear genes, nuclear-encoded genes, splicing occurs in the cell nucleus, nucleus either during or immediately after Transcription (biology), transcription. For those eukaryotic transcription, eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translation (biology), translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5' Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. The Cap-0 is the base cap structure, however, the first and second transcribed nucleotides can also be 2' O-methylated, leading to the Cap-1 and Cap-2 structures, respectively. This is more common in higher eukaryotes an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics ( phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA repair#DNA damage, DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30 nm fiber. ''Current opinion in cell biolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Embryo
An embryo ( ) is the initial stage of development for a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm cell. The resulting fusion of these two cells produces a single-celled zygote that undergoes many cell divisions that produce cells known as blastomeres. The blastomeres (4-cell stage) are arranged as a solid ball that when reaching a certain size, called a morula, (16-cell stage) takes in fluid to create a cavity called a blastocoel. The structure is then termed a blastula, or a blastocyst in mammals. The mammalian blastocyst hatches before implantating into the endometrial lining of the womb. Once implanted the embryo will continue its development through the next stages of gastrulation, neurulation, and organogenesis. Gastrulation is the formation of the three germ layers that will form all of the different parts of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microinjection
Microinjection is the use of a glass micropipette to inject a liquid substance at a microscopic or borderline macroscopic level. The target is often a living cell but may also include intercellular space. Microinjection is a simple mechanical process usually involving an inverted microscope with a Optical power, magnification power of around 200x (though sometimes it is performed using a dissecting stereo microscope at 40–50x or a traditional Optical microscope#Compound microscope, compound upright microscope at similar power to an inverted model). For processes such as cellular or pronucleus, pronuclear injection the target cell is positioned under the microscope and two micromanipulation, micromanipulators—one holding the pipette and one holding a microcapillary needle usually between 0.5 and 5 μm in diameter (larger if injecting stem cells into an embryo)—are used to penetrate the cell membrane and/or the nuclear envelope. In this way the process can be used to int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
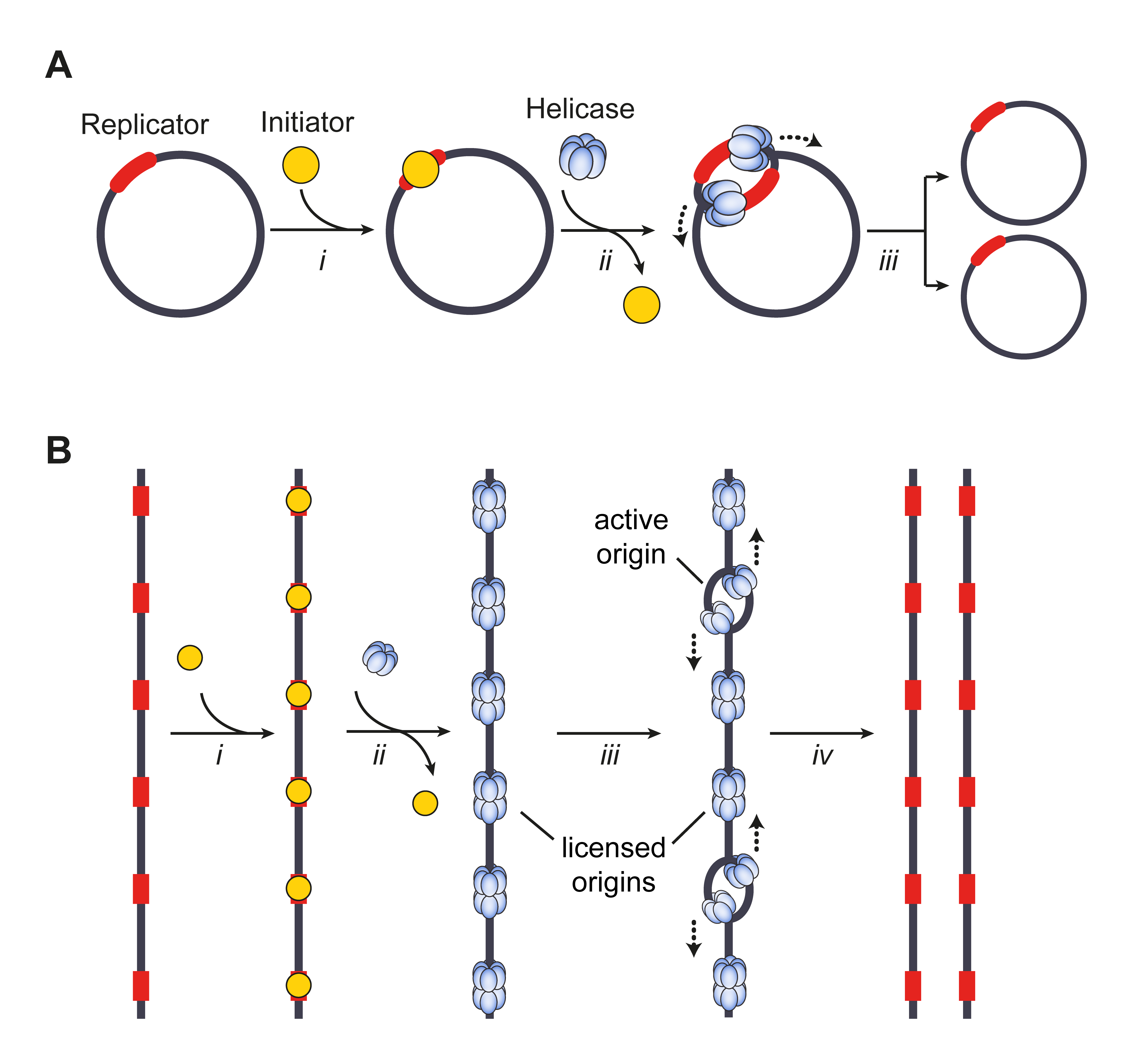
Origin Of Replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. Material was copied from this source, which is available under Creative Commons Attribution 4.0 International License This can either involve the DNA replication, replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA virus, DNA or RNA virus, RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibiotic Resistance
Antimicrobial resistance (AMR or AR) occurs when microbes evolve mechanisms that protect them from antimicrobials, which are drugs used to treat infections. This resistance affects all classes of microbes, including bacteria (antibiotic resistance), viruses (antiviral resistance), parasites (antiparasitic resistance), and fungi (antifungal resistance). Together, these adaptations fall under the AMR umbrella, posing significant challenges to healthcare worldwide. Misuse and improper management of antimicrobials are primary drivers of this resistance, though it can also occur naturally through genetic mutations and the spread of resistant genes. Antibiotic resistance, a significant AMR subset, enables bacteria to survive antibiotic treatment, complicating infection management and treatment options. Resistance arises through spontaneous mutation, horizontal gene transfer, and increased selective pressure from antibiotic overuse, both in medicine and agriculture, which accelerat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selectable Marker
A selectable marker is a gene introduced into cell (biology), cells, especially bacteria or cells in cell culture, culture, which confers one or more traits suitable for artificial selection. They are a type of reporter gene used in laboratory microbiology, molecular biology, and genetic engineering to indicate the success of a transfection or genetic transformation, transformation or other procedure meant to introduce foreign DNA into a cell. Selectable markers are often antibiotic resistance genes: bacteria subjected to a procedure by which exogenous DNA containing an antibiotic resistance gene (usually alongside other genes of interest) has been introduced are grown on a medium containing an antibiotic, such that only those bacterial cells which have successfully taken up and gene expression, expressed the introduced genetic material, including the gene which confers antibiotic resistance, can survive and produce colony (biology), colonies. The genes encoding resistance to antibio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |