|
P53 Protein
p53, also known as tumor protein p53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory transcription factor protein that is often mutated in human cancers. The p53 proteins (originally thought to be, and often spoken of as, a single protein) are crucial in vertebrates, where they prevent cancer formation. As such, p53 has been described as "the guardian of the genome" because of its role in conserving stability by preventing genome mutation. Hence ''TP53'' ''italics'' are used to denote the ''TP53'' gene name and distinguish it from the protein it encodes is classified as a tumor suppressor gene. The ''TP53'' gene is the most frequently mutated gene (>50%) in human cancer, indicating that the ''TP53'' gene plays a crucial role in preventing cancer formation. ''TP53'' gene encodes proteins that bind to DNA and regulate gene expression to prevent mutations of the genome. In addition to the full-length protein, the human ''TP53 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, USA. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the George ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG). Proline is the only proteinogenic amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring. History and etymology Proline was first isolated in 1900 by Richard Willstätter who obtained the amino a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LMO3
LIM domain only protein 3 is a transcription co-factor, which in humans is encoded by the ''LMO3'' gene. LMO3 interacts with the tumor suppressor p53 and regulates its function. LMO3 is considered to be an oncogene in Neuroblastoma Neuroblastoma (NB) is a type of cancer that forms in certain types of nerve tissue. It most frequently starts from one of the adrenal glands but can also develop in the head, neck, chest, abdomen, or Vertebral column, spine. Symptoms may include .... References Further reading * * * * * * {{gene-12-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
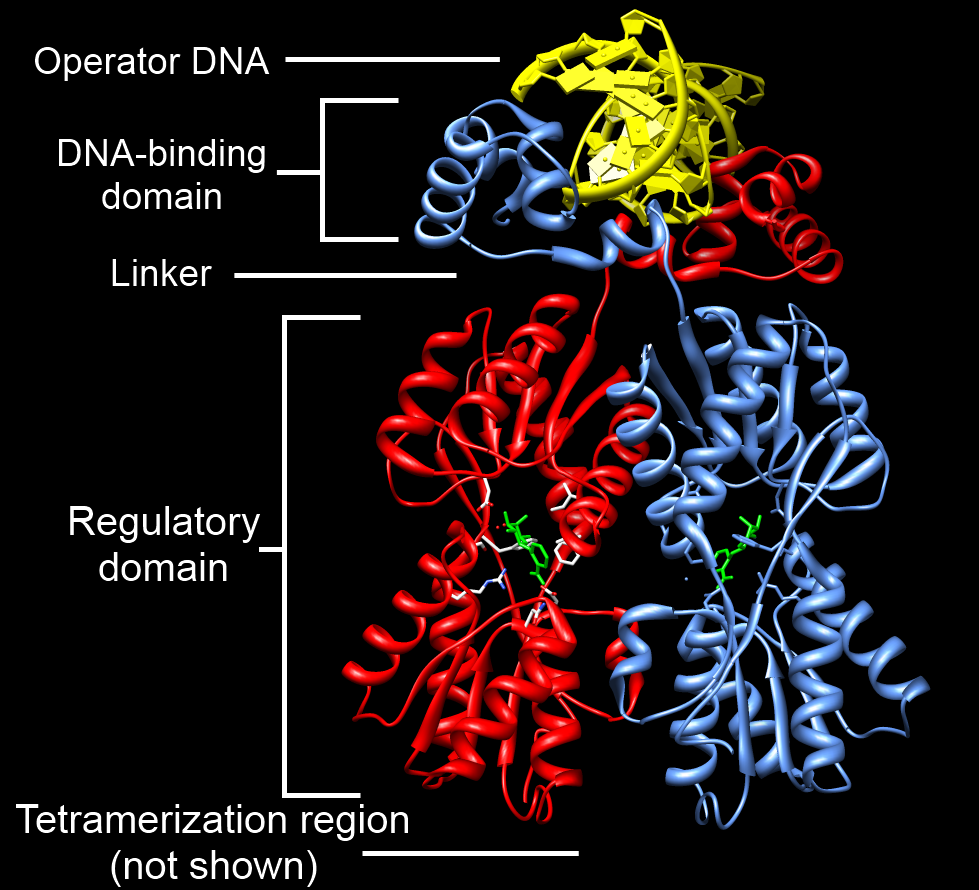
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAPK
A mitogen-activated protein kinase (MAPK or MAP kinase) is a type of serine/threonine-specific protein kinases involved in directing cellular responses to a diverse array of stimuli, such as mitogens, osmotic stress, heat shock and proinflammatory cytokines. They regulate cell functions including proliferation, gene expression, differentiation, mitosis, cell survival, and apoptosis. MAP kinases are found in eukaryotes only, but they are fairly diverse and encountered in all animals, fungi and plants, and even in an array of unicellular eukaryotes. MAPKs belong to the CMGC (CDK/MAPK/GSK3/CLK) kinase group. The closest relatives of MAPKs are the cyclin-dependent kinases (CDKs). Discovery The first mitogen-activated protein kinase to be discovered was ERK1 ( MAPK3) in mammals. Since ERK1 and its close relative ERK2 ( MAPK1) are both involved in growth factor signaling, the family was termed "mitogen-activated". With the discovery of other members, even from distant organis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biology), morphology) and death. These changes include Bleb (cell biology), blebbing, Plasmolysis, cell shrinkage, Karyorrhexis, nuclear fragmentation, Pyknosis, chromatin condensation, Apoptotic DNA fragmentation, DNA fragmentation, and mRNA decay. The average adult human loses 50 to 70 1,000,000,000, billion cells each day due to apoptosis. For the average human child between 8 and 14 years old, each day the approximate loss is 20 to 30 billion cells. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transactivation Domain
The transactivation domain or trans-activating domain (TAD) is a transcription factor scaffold domain which contains binding sites for other proteins such as transcription coregulators. These binding sites are frequently referred to as activation functions (AFs). TADs are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4 transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic, respectively. In general we can distinguish four classes of TADs: * acidic domains (called also “acid blobs” or “negative noodles", rich in D and E amino acids, present in Gal4, Gcn4 and VP16). * glutamine-rich domains (contains multiple repetitions like "QQQXXXQQQ", present in SP1) * proline-rich domains (contains repetitions like "PPPXXXPPP" present i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain (protein)
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer af ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P53 Schematic
p53, also known as tumor protein p53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory transcription factor protein that is often mutated in human cancers. The p53 proteins (originally thought to be, and often spoken of as, a single protein) are crucial in vertebrates, where they prevent cancer formation. As such, p53 has been described as "the guardian of the genome" because of its role in conserving stability by preventing genome mutation. Hence ''TP53''Gene nomenclature#Vertebrate gene and protein symbol conventions, ''italics'' are used to denote the ''TP53'' gene name and distinguish it from the protein it encodes is classified as a tumor suppressor gene. The ''TP53'' gene is the most frequently mutated gene (>50%) in human cancer, indicating that the ''TP53'' gene plays a crucial role in preventing cancer formation. ''TP53'' gene encodes proteins that bind to DNA and regulate gene expression to prevent mutations of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |