|
List Of RNA-Seq Bioinformatics Tools
RNA-Seq is a technique that allows transcriptome studies (see also Transcriptomics technologies) based on next-generation sequencing technologies. This technique is largely dependent on bioinformatics tools developed to support the different steps of the process. Here are listed some of the principal tools commonly employed and links to some important web resources. Design Design is a fundamental step of a particular RNA-Seq experiment. Some important questions like sequencing depth/coverage or how many biological or technical replicates must be carefully considered. Design review. *PROPER: PROspective Power Evaluation for RNAseq. *RNAtor: an Android Application to calculate optimal parameters for popular tools and kits available for DNA sequencing projects. *Scotty: a web tool for designing RNA-Seq experiments to measure differential gene expression. *ssizeRNA Sample Size Calculation for RNA-Seq Experimental Design. Quality control, trimming, error correction and pre-processing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/ SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Burrows–Wheeler Transform
The Burrows–Wheeler transform (BWT) rearranges a character string into runs of similar characters, in a manner that can be reversed to recover the original string. Since compression techniques such as move-to-front transform and run-length encoding are more effective when such runs are present, the BWT can be used as a preparatory step to improve the efficiency of a compression algorithm, and is used this way in software such as bzip2. The algorithm can be implemented efficiently using a suffix array thus reaching linear time complexity. It was invented by David Wheeler in 1983, and later published by him and Michael Burrows in 1994. Their paper included a compression algorithm, called the Block-sorting Lossless Data Compression Algorithm or BSLDCA, that compresses data by using the BWT followed by move-to-front coding and Huffman coding or arithmetic coding. Description The transform is done by constructing a matrix (known as the Burrows-Wheeler Matrix) whose rows are the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bisulfite Sequencing
Bisulfite sequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity. Treatment of DNA with bisulfite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Therefore, DNA that has been treated with bisulfite retains only methylated cytosines. Thus, bisulfite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues, yielding single-nucleotide resolution information about the methylation status of a segment of DNA. Various analyses can be performed on the altered sequence to retrieve this informatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hidden Markov Model
A hidden Markov model (HMM) is a Markov model in which the observations are dependent on a latent (or ''hidden'') Markov process (referred to as X). An HMM requires that there be an observable process Y whose outcomes depend on the outcomes of X in a known way. Since X cannot be observed directly, the goal is to learn about state of X by observing Y. By definition of being a Markov model, an HMM has an additional requirement that the outcome of Y at time t = t_0 must be "influenced" exclusively by the outcome of X at t = t_0 and that the outcomes of X and Y at t < t_0 must be conditionally independent of at given at time . Estimation of the parameters in an HMM can be performed using maximum likelihood estimation. For linear chain HMMs, the Baum–Welch algorithm can be used to estimate parameters. Hidden Markov models are known for their applications to thermodynamics, statistical mechanics, physics, chem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Applied Biosystems
Applied Biosystems is one of various brands under the Life Technologies brand of Thermo Fisher Scientific corporation. The brand is focused on integrated systems for genetic analysis, which include computerized machines and the consumables used within them (such as reagents). In 2008, a merger between Applied Biosystems and Invitrogen was finalized, creating Life Technologies. The latter was acquired by Thermo Fisher Scientific in 2014. Prior to 2008, the Applied Biosystems brand was owned by various entities in a corporate group parented by PerkinElmer. The roots of Applied Biosystems trace back to GeneCo (Genetic Systems Company), a pioneer biotechnology company founded in 1981 in Foster City, California.Applied Biosystems Timeline , AppliedBiosystems.com Through the 1980s and early 19 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expressed Sequence Tags
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and were instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases (e.g. GenBank 1 January 2013, all species). EST approaches have largely been superseded by whole genome and transcriptome sequencing and metagenome sequencing. An EST results from one-shot sequencing of a cloned cDNA. The cDNAs used for EST generation are typically individual clones from a cDNA library. The resulting sequence is a relatively low-quality fragment whose length is limited by current technology to approximately 500 to 800 nucleotides. Because these clones consist of DNA that is complementary to mRNA, the ESTs represent portions of expressed genes. They may be represented in databases as either cDNA/mRNA sequence or as the reverse complement o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Q-gram
An ''n''-gram is a sequence of ''n'' adjacent symbols in particular order. The symbols may be ''n'' adjacent letters (including punctuation marks and blanks), syllables, or rarely whole words found in a language dataset; or adjacent phonemes extracted from a speech-recording dataset, or adjacent base pairs extracted from a genome. They are collected from a text corpus or speech corpus. If Latin numerical prefixes are used, then ''n''-gram of size 1 is called a "unigram", size 2 a "bigram" (or, less commonly, a "digram") etc. If, instead of the Latin ones, the English cardinal numbers are furtherly used, then they are called "four-gram", "five-gram", etc. Similarly, using Greek numerical prefixes such as "monomer", "dimer", "trimer", "tetramer", "pentamer", etc., or English cardinal numbers, "one-mer", "two-mer", "three-mer", etc. are used in computational biology, for polymers or oligomers of a known size, called ''k''-mers. When the items are words, -grams may also be ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Picard MarkDuplicates
Picard may refer to: Places * Picard, Quebec, Canada * Picard, California, United States * Picard (crater), a lunar impact crater in Mare Crisium People and fictional characters *Picard (name), a list of people and fictional characters with the surname * Picards, a religious sect in the fifteenth century Other uses *'' Star Trek: Picard'', a television series focusing on the character of Jean-Luc Picard *Picard language, a language spoken in France and Belgium *Picard (satellite), an orbiting solar observatory built by CNES *Picard (grape), an alternative name for several wine grape varieties *''TSS Duke of Cumberland'' or ''Picard'', a steamship that operated between Tilbury and Dunkirk from 1927 to 1936 *Picard Surgelés, French retailer of frozen foods See also * * *Berger Picard, French breed of dog of the herding group of breeds *Les Fatals Picards, a French band *MusicBrainz Picard, a cross platform tagger for MusicBrainz database *Picard group, a mathematical object descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MapReduce
MapReduce is a programming model and an associated implementation for processing and generating big data sets with a parallel and distributed algorithm on a cluster. A MapReduce program is composed of a ''map'' procedure, which performs filtering and sorting (such as sorting students by first name into queues, one queue for each name), and a '' reduce'' method, which performs a summary operation (such as counting the number of students in each queue, yielding name frequencies). The "MapReduce System" (also called "infrastructure" or "framework") orchestrates the processing by marshalling the distributed servers, running the various tasks in parallel, managing all communications and data transfers between the various parts of the system, and providing for redundancy and fault tolerance. The model is a specialization of the ''split-apply-combine'' strategy for data analysis. It is inspired by the map and reduce functions commonly used in functional programming,"Our abstracti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Needleman–Wunsch Algorithm
The Needleman–Wunsch algorithm is an algorithm used in bioinformatics to align protein or nucleotide sequences. It was one of the first applications of dynamic programming to compare biological sequences. The algorithm was developed by Saul B. Needleman and Christian D. Wunsch and published in 1970. The algorithm essentially divides a large problem (e.g. the full sequence) into a series of smaller problems, and it uses the solutions to the smaller problems to find an optimal solution to the larger problem. It is also sometimes referred to as the optimal matching algorithm and the global alignment technique. The Needleman–Wunsch algorithm is still widely used for optimal global alignment, particularly when the quality of the global alignment is of the utmost importance. The algorithm assigns a score to every possible alignment, and the purpose of the algorithm is to find all possible alignments having the highest score. Introduction This algorithm can be used for any two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FM-index
In computer science, an FM-index is a compressed full-text substring index based on the Burrows–Wheeler transform, with some similarities to the suffix array. It was created by Paolo Ferragina and Giovanni Manzini,Paolo Ferragina and Giovanni Manzini (2000)"Opportunistic Data Structures with Applications".Proceedings of the 41st Annual Symposium on Foundations of Computer Science. p.390. who describe it as an opportunistic data structure as it allows compression of the input text while still permitting fast substring queries. The name stands for Full-text index in Minute space. It can be used to efficiently find the number of occurrences of a pattern within the compressed text, as well as locate the position of each occurrence. The query time, as well as the required storage space, has a sublinear complexity with respect to the size of the input data. The original authors have devised improvements to their original approach and dubbed it "FM-Index version 2". A further improvem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
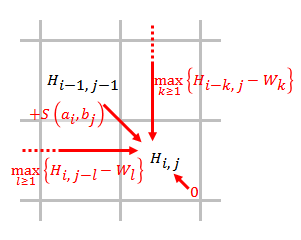
Smith–Waterman Algorithm
The Smith–Waterman algorithm performs local sequence alignment; that is, for determining similar regions between two strings of nucleic acid sequences or protein sequences. Instead of looking at the entire sequence, the Smith–Waterman algorithm compares segments of all possible lengths and optimizes the similarity measure. The algorithm was first proposed by Temple F. Smith and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix and the gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm is that negative scoring matrix cells are set to zero. Traceback procedure starts at the highest scoring matrix cell and proceeds until a cell with score zero is encountered, yielding the h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |