|
LTR Retrotransposons
LTR retrotransposons are class I transposable elements (TEs) characterized by the presence of long terminal repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse transcription of their mRNA and integration of the newly created cDNA into another genomic location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that the rate of horizontal transfer in LTR-retrotransposons is much lower than the vertical transfer by passing active TE insertions to the progeny. LTR retrotransposons that form virus-like particles are classified under ''Ortervirales''. Their size ranges from a few hundred base pairs to 30 kb, the largest species reported to date are members of the Burro retrotransposon family in ''Schmidtea mediterranea''. In plant genomes, LTR retrotransposons are the major repetitive sequence class constituting more than 75% of the maize genome. LTR retrotransposons make up about 8% of the h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LTR Retrotransposon
LTR retrotransposons are class I transposable elements (TEs) characterized by the presence of long terminal repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse transcription of their mRNA and integration of the newly created cDNA into another genomic location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that the rate of horizontal transfer in LTR-retrotransposons is much lower than the vertical transfer by passing active TE insertions to the progeny. LTR retrotransposons that form virus-like particles are classified under ''Ortervirales''. Their size ranges from a few hundred base pairs to 30 kb, the largest species reported to date are members of the Burro retrotransposon family in '' Schmidtea mediterranea''. In plant genomes, LTR retrotransposons are the major repetitive sequence class constituting more than 75% of the maize genome. LTR retrotransposons make up about 8% of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pol (HIV)
Pol (DNA polymerase) refers to a gene in retroviruses, or the protein produced by that gene. Products of pol include: * Reverse transcriptase (RT), which transcribes the viral RNA into double-stranded DNA. * Integrase, which integrates the DNA produced by RT into the host cell's genome. * Protease A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalysis, catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products ..., an enzyme that cuts proteins into segments. HIV's ''gag'' and ''pol'' genes do not encode their proteins in their final form but as larger polyproteins that the HIV protease cleaves into separate functional units. See also * Gag/pol translational readthrough site References External links * * {{Viral proteins Viral structural proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gymnosperms
The gymnosperms ( ; ) are a group of woody, perennial Seed plant, seed-producing plants, typically lacking the protective outer covering which surrounds the seeds in flowering plants, that include Pinophyta, conifers, cycads, Ginkgo, and gnetophyta, gnetophytes, forming the clade Gymnospermae. The term ''gymnosperm'' comes from the composite word in ( and ), and literally means 'naked seeds'. The name is based on the unenclosed condition of their seeds (called ovules in their unfertilized state). The non-encased condition of their seeds contrasts with the seeds and ovules of flowering plants (angiosperms), which are enclosed within an Ovary (botany), ovary. Gymnosperm seeds develop either on the surface of scales or Leaf, leaves, which are often modified to form Conifer cone, cones, or on their own as in Taxus, yew, ''Torreya'', and ''Ginkgo''. The life cycle of a gymnosperm involves alternation of generations, with a dominant diploid sporophyte phase, and a reduced haploid gam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bryophytes
Bryophytes () are a group of land plants ( embryophytes), sometimes treated as a taxonomic division referred to as Bryophyta '' sensu lato'', that contains three groups of non-vascular land plants: the liverworts, hornworts, and mosses. In the strict sense, the division Bryophyta consists of the mosses only. Bryophytes are characteristically limited in size and prefer moist habitats although some species can survive in drier environments. The bryophytes consist of about 20,000 plant species. Bryophytes produce enclosed reproductive structures ( gametangia and sporangia), but they do not produce flowers or seeds. They reproduce sexually by spores and asexually by fragmentation or the production of gemmae. Though bryophytes were considered a paraphyletic group in recent years, almost all of the most recent phylogenetic evidence supports the monophyly of this group, as originally classified by Wilhelm Schimper in 1879. The term ''bryophyte'' comes . Features The defining ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Algae
Algae ( , ; : alga ) is an informal term for any organisms of a large and diverse group of photosynthesis, photosynthetic organisms that are not plants, and includes species from multiple distinct clades. Such organisms range from unicellular microalgae, such as cyanobacteria, ''Chlorella'', and diatoms, to multicellular macroalgae such as kelp or brown algae which may grow up to in length. Most algae are aquatic organisms and lack many of the distinct cell and tissue types, such as stomata, xylem, and phloem that are found in embryophyte, land plants. The largest and most complex marine algae are called seaweeds. In contrast, the most complex freshwater forms are the Charophyta, a Division (taxonomy), division of green algae which includes, for example, ''Spirogyra'' and stoneworts. Algae that are carried passively by water are plankton, specifically phytoplankton. Algae constitute a Polyphyly, polyphyletic group because they do not include a common ancestor, and although Eu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNAs
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code. Overview The process of translation starts with the information stored in the nucleotide sequence of DNA. This is first transformed into mRNA, then tRNA specifies which three-nucleotide codon from the genetic code corresponds to which amino acid. Each mRNA codon is recognized by a particular type of tRNA, which docks to it along a t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
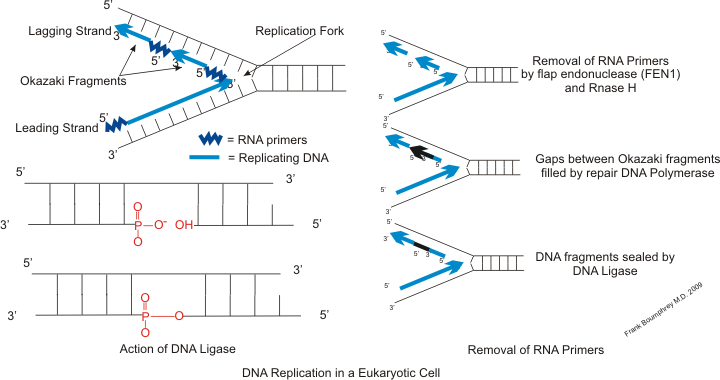
Primer Binding Site
A primer binding site is a region of a nucleotide sequence where an RNA or DNA single-stranded primer binds to start replication. The primer binding site is on one of the two complementary strands of a double-stranded nucleotide polymer, in the strand that is to be copied, or is within a single-stranded nucleotide polymer sequence. DNA Replication DNA replication is the semi-conservative biological process of two DNA strands copying themselves, resulting in two identical copies of DNA. This process is considered semi-conservative because, after replication, each copy of DNA contains a strand from the original DNA molecule and a strand from the newly-synthesized DNA molecule. An RNA primer is a short chain of single-stranded RNA, consisting of roughly five to ten nucleotides complementary to the DNA template strand. DNA polymerase will then take each nucleotide and make a new complementary DNA strand to the template strand, but only in the 5' to 3' direction. One of the new st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcription
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B virus, hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryote, eukaryotic cells to extend the telomeres at the ends of their Chromosome#Eukaryotes, linear chromosomes. The process does not violate the flows of genetic information as described by the classical central dogma of molecular biology, central dogma, but rather expands it to include transfers of information from RNA to DNA. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then inte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA polymerase, RNAP enzymes found in the nucleus of eukaryote, eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoter (biology), promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, biochemists Robert G. Roeder and William J. Rutter, William Rutter discovered there are total three distinct nuclear RNA polymerases, an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integrase
Retroviral integrase (IN) is an enzyme An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ... produced by a retrovirus (such as HIV) that integrates (forms covalent links between) its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases ( recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination. The macromolecular complex of an IN macromolecule bound to the ends of the viral DNA ends has been referred to as the '' intasome''; IN is a key component in this and the retroviral pre-integration complex. Structure All retroviral IN proteins contain three canonical domains, connected by flexible linkers: * an N-terminal HH-CC zinc-binding domain (a three-heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |