|
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics. History The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm. FASTA is pronounced "f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA Format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format allows for sequence names and comments to precede the sequences. It originated from the FASTA software package and has since become a near-universal standard in bioinformatics. The simplicity of FASTA format makes it easy to manipulate and parse sequences using text-processing tools and scripting languages. Overview A sequence begins with a greater-than character (">") followed by a description of the sequence (all in a single line). The lines immediately following the description line are the sequence representation, with one letter per amino acid or nucleic acid, and are typically no more than 80 characters in length. For example: >MCHU - Calmodulin - Human, rabbit, bovine, rat, and chicken MADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTV ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
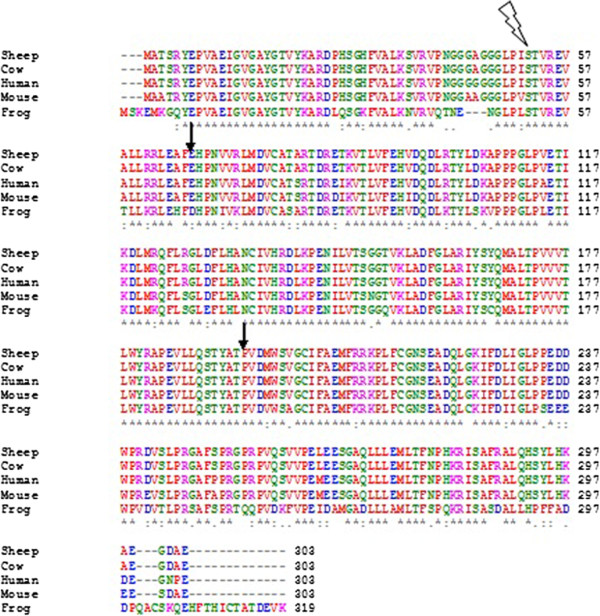
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix (mathematics), matrix. Gaps are inserted between the Residue (chemistry), residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences such as calculating the Edit distance, distance cost between strings in a natural language, or to display financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence ali ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST (biotechnology)
In bioinformatics, BLAST (basic local alignment search tool) is an algorithm and program for comparing Primary structure, primary biological sequence information, such as the amino acid, amino-acid sequences of proteins or the nucleotides of DNA sequence, DNA and/or RNA sequences. A BLAST search enables a researcher to compare a subject protein or nucleotide sequence (called a query) with a library or database of sequences, and identify database sequences that resemble the query sequence above a certain threshold. For example, following the discovery of a previously unknown gene in the Mus musculus, mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. Background BLAST is one of the most widely used bioinformatics programs for sequence searching. It addresses a fundamental problem in bioinformatics research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
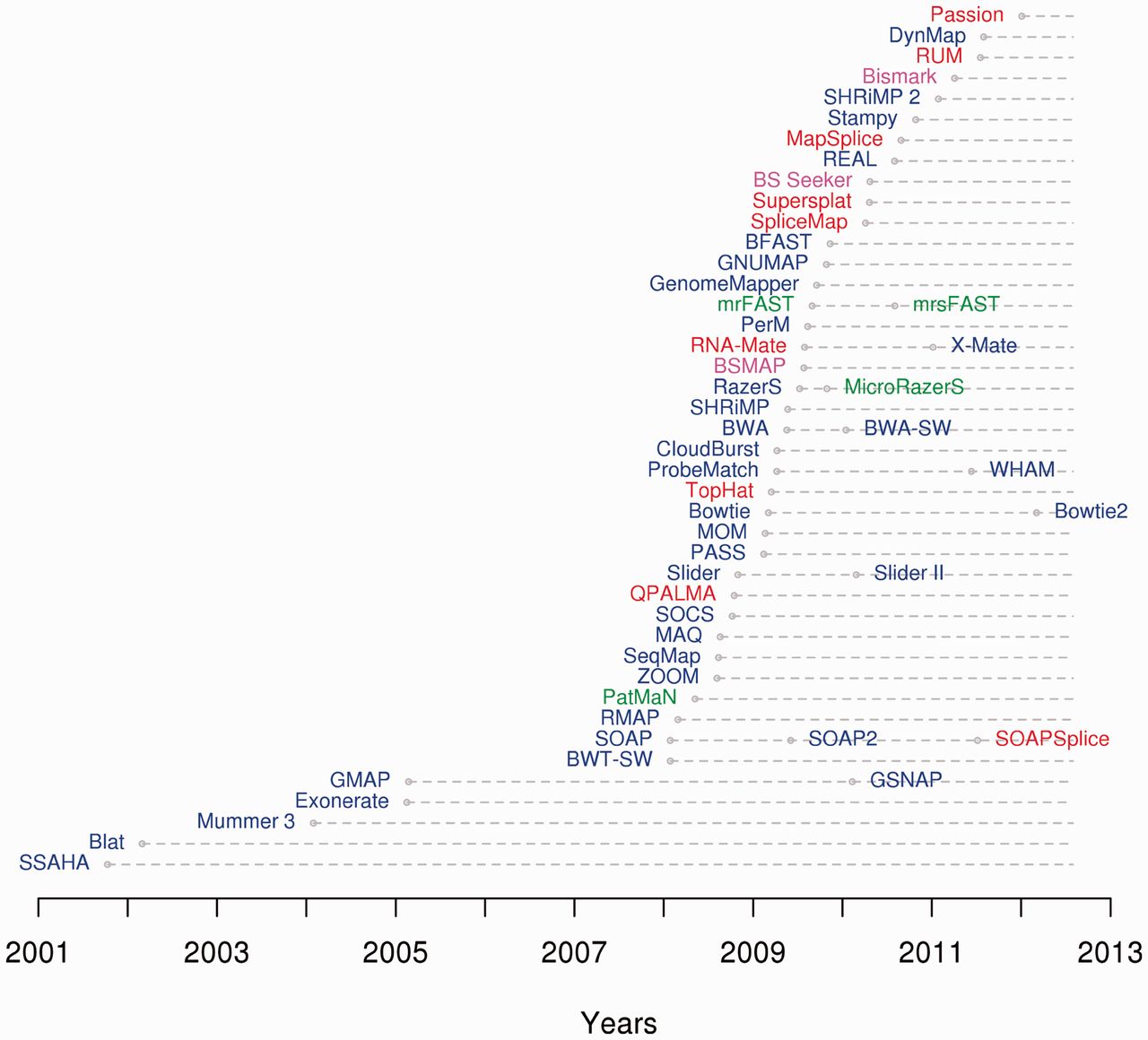
Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Seq Sequence In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
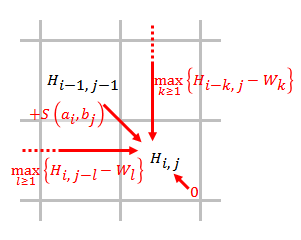
Smith–Waterman Algorithm
The Smith–Waterman algorithm performs local sequence alignment; that is, for determining similar regions between two strings of nucleic acid sequences or protein sequences. Instead of looking at the entire sequence, the Smith–Waterman algorithm compares segments of all possible lengths and optimizes the similarity measure. The algorithm was first proposed by Temple F. Smith and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix and the gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm is that negative scoring matrix cells are set to zero. Traceback procedure starts at the highest scoring matrix cell and proceeds until a cell with score zero is encountered, yielding the h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T-Coffee
T-Coffee (Tree-based Consistency Objective Function for Alignment Evaluation) is a multiple sequence alignment software using a progressive approach. It generates a library of pairwise alignments to guide the multiple sequence alignment. It can also combine multiple sequences alignments obtained previously and in the latest versions can use structural information from Protein Data Bank (PDB) files (3D-Coffee). It has advanced features to evaluate the quality of the alignments and some capacity for identifying occurrence of motifs (Mocca). It produces alignment in the aln format (Clustal) by default, but can also produce PIR, MSF, and FASTA format. The most common input formats are supported (FASTA, Protein Information Resource (PIR)). Algorithm T-Coffee algorithm consist of two main features, the first by, using heterogeneous data sources, can provide simple and flexible means to generate multiple alignments. T-coffee can compute multiple alignments using a library that was generat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
William Pearson (scientist)
William Raymond Pearson is professor of biochemistry and molecular Genetics in the School of Medicine at the University of Virginia. Pearson is best known for the development of the FASTA format. Education Pearson graduated with a BS in chemistry from the University of Illinois Urbana-Champaign. He received his PhD in 1977 from Caltech. As a graduate student, he published several papers describing computer programs for analyzing biological data. Career and research After his PhD, Pearson did a postdoctoral fellowship at Johns Hopkins University. In 1983, he joined the faculty of Biochemistry at the University of Virginia School of Medicine. Immediately after joining the faculty, he collaborated with David J. Lipman at the NIH to write the FASTP program, and later FASTA. Pearson's research interests are in computational biology. He was named an American Association for the Advancement of Science (AAAS) Fellow in 2008, and an International Society for Computational Biology (ISCB) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clustal
Clustal is a computer program used for multiple sequence alignment in bioinformatics. The software and its algorithms have gone through several iterations, with ClustalΩ (Omega) being the latest version . It is available as standalone software, via a Clustal#External links, web interface, and through a server hosted by the European Bioinformatics Institute. Clustal has been an important bioinformatic software, with two of its academic publications amongst the top 100 papers cited of all time, according to Nature (journal), Nature in 2014. History Version history * Clustal: The original software for multiple sequence alignments, created by Desmond G. Higgins, Des Higgins in 1988, was based on deriving a guide tree from pairwise sequences of Amino acid, amino acids or Nucleotide, nucleotides. * ClustalV: The second generation of Clustal, released in 1992. It introduced the ability to create new alignments from existing alignments in a process known as phylogenetic tree re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
European Bioinformatics Institute
The European Bioinformatics Institute (EMBL-EBI) is an intergovernmental organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wellcome Genome Campus in Hinxton near Cambridge, and employs over 600 full-time equivalent (FTE) staff. Further, the EMBL-EBI hosts training programs that teach scientists the fundamentals of the work with biological data and promote the plethora of bioinformatic tools available for their research, both EMBL-EBI-based and not so. Bioinformatic services One of the roles of the EMBL-EBI is to index and maintain biological data in a set of databases, including Ensembl (housing whole genome sequence data), UniProt (protein sequence and annotation database) and Protein Data Bank (protein and nucleic acid tertiary structure database). A variety of online services and tools is provided, such as Basic Local Alignment Search Tool (BLAST) or Clust ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substitution Matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a Nucleic acid sequence, nucleotide sequence or a Protein primary structure, protein sequence changes to other character states over evolutionary time. The information is often in the form of Logit, log odds of finding two specific character states aligned and depends on the assumed number of evolutionary changes or sequence dissimilarity between compared sequences. It is an application of a stochastic matrix. Substitution matrices are usually seen in the context of amino acid or DNA sequence alignments, where they are used to calculate similarity scores between the aligned sequences. Background In the process of evolution, from one generation to the next the amino acid sequences of an organism's proteins are gradually altered through the action of DNA mutations. For example, the sequence ALEIRYLRD could mutate into the sequence ALEINYLRD in one step, and pos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Similarity Measure
In statistics and related fields, a similarity measure or similarity function or similarity metric is a real-valued function that quantifies the similarity between two objects. Although no single definition of a similarity exists, usually such measures are in some sense the inverse of distance metrics: they take on large values for similar objects and either zero or a negative value for very dissimilar objects. Though, in more broad terms, a similarity function may also satisfy metric axioms. Cosine similarity is a commonly used similarity measure for real-valued vectors, used in (among other fields) information retrieval to score the similarity of documents in the vector space model. In machine learning, common kernel functions such as the RBF kernel can be viewed as similarity functions. Use of different similarity measure formulas Different types of similarity measures exist for various types of objects, depending on the objects being compared. For each type of object there ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |