|
Dihydrofolate Reductase
Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor, which can be converted to the kinds of tetrahydrofolate cofactors used in one-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the ''DHFR'' gene. It is found in the q14.1 region of chromosome 5. There are two structural classes of DHFR, evolutionarily unrelated to each other. The former is usually just called DHFR and is found in bacterial chromosomes and animals. In bacteria, however, antibiotic pressure has caused this class to evolve different patterns of binding diaminoheterocyclic molecules, leading to many "types" named under this class, while mammalian ones remain highly similar. The latter (type II), represented by the plastid-encoded R67, is a tiny enzyme that works by forming a homotetramer. Function Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a proton shuttle required for the de no ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrobiopterin
Dihydrobiopterin (BH2) is a pteridine compound produced in the synthesis of L-DOPA, dopamine, serotonin, norepinephrine and epinephrine. It is restored to the required cofactor tetrahydrobiopterin by dihydrobiopterin reductase. See also * Pteridine * Tetrahydrobiopterin Tetrahydrobiopterin (BH4, THB), also known as sapropterin ( INN), is a cofactor of the three aromatic amino acid hydroxylase enzymes, used in the metabolism of amino acid phenylalanine and in the biosynthesis of the neurotransmitters serotoni ... * Dihydropteridine reductase deficiency Biomolecules {{biochem-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
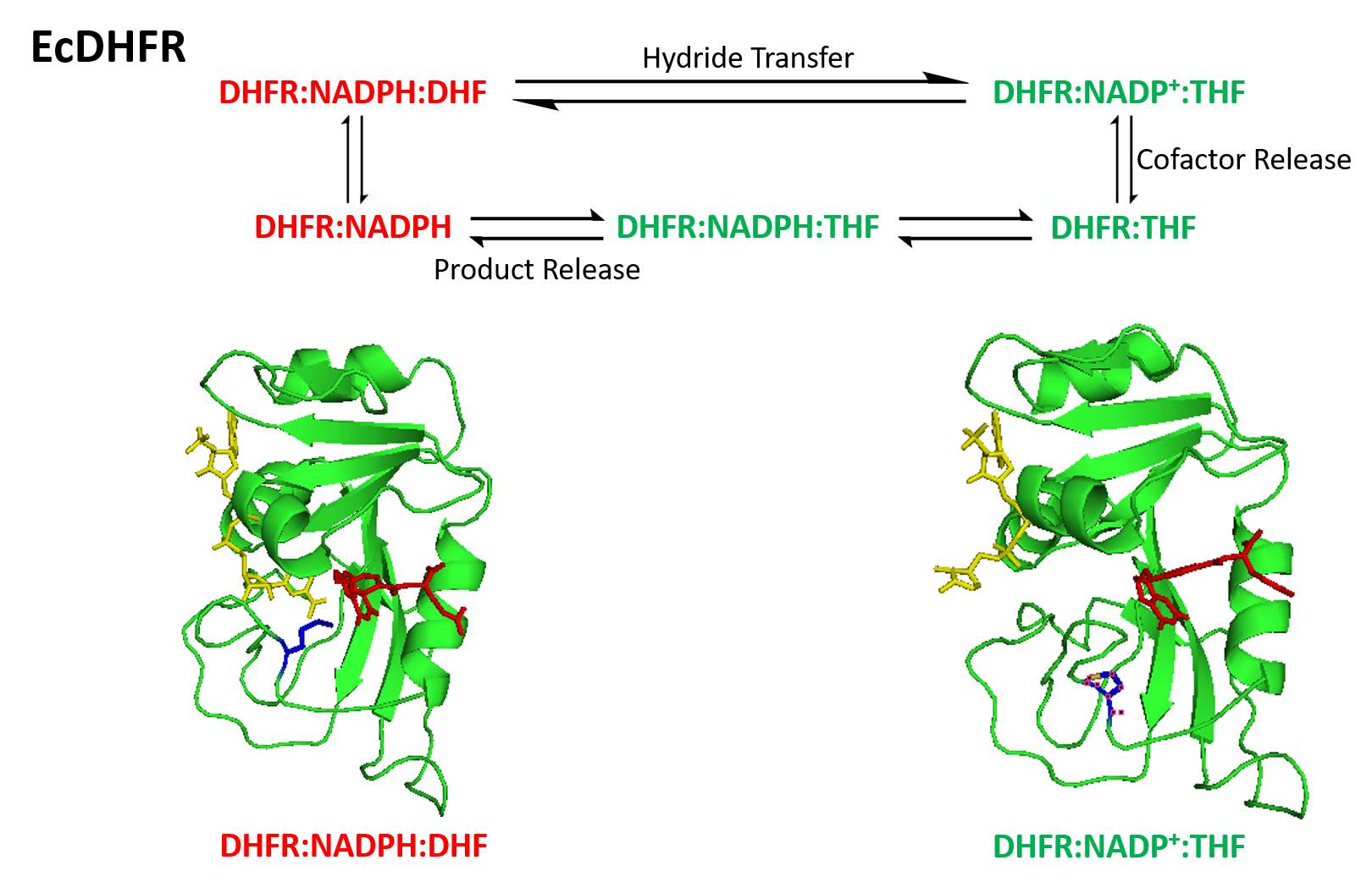
DHFR + NADPH + Folate (Met20 Loop)
Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor, which can be converted to the kinds of tetrahydrofolate cofactor (biochemistry), cofactors used in one-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the ''DHFR'' gene. It is found in the q14.1 region of chromosome 5. There are two structural classes of DHFR, evolutionarily unrelated to each other. The former is usually just called DHFR and is found in bacterial chromosomes and animals. In bacteria, however, antibiotic pressure has caused this class to evolve different patterns of binding diaminoheterocyclic molecules, leading to many "types" named under this class, while mammalian ones remain highly similar. The latter (type II), represented by the plastid-encoded R67, is a tiny enzyme that works by forming a homotetramer. Function Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a proton shuttle re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADP+
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form, whereas NADP is the oxidized form. NADP is used by all forms of cellular life. NADP is essential for life because it is needed for cellular respiration. NADP differs from NAD by the presence of an additional phosphate group on the 2' position of the ribose ring that carries the adenine moiety. This extra phosphate is added by NAD+ kinase and removed by NADP+ phosphatase. Biosynthesis NADP In general, NADP+ is synthesized before NADPH is. Such a reaction usually starts with NAD+ from either the de-novo or the salvage pathway, with NAD+ kinase adding the extra phosphate group. ADP-ribosyl cyclase allows for synthesis from nicotinamide in the salvage pathway, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DHFR Reaction Scheme
Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor, which can be converted to the kinds of tetrahydrofolate cofactors used in one-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the ''DHFR'' gene. It is found in the q14.1 region of chromosome 5. There are two structural classes of DHFR, evolutionarily unrelated to each other. The former is usually just called DHFR and is found in bacterial chromosomes and animals. In bacteria, however, antibiotic pressure has caused this class to evolve different patterns of binding diaminoheterocyclic molecules, leading to many "types" named under this class, while mammalian ones remain highly similar. The latter (type II), represented by the plastid-encoded R67, is a tiny enzyme that works by forming a homotetramer. Function Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a proton shuttle required for the de novo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Substrate
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed. In the former sense, a reagent is added to the ''substrate'' to generate a product through a chemical reaction. The term is used in a similar sense in synthetic and organic chemistry, where the substrate is the chemical of interest that is being modified. In biochemistry, an enzyme substrate is the material upon which an enzyme acts. When referring to Le Chatelier's principle, the substrate is the reagent whose concentration is changed. ;Spontaneous reaction : :*Where S is substrate and P is product. ;Catalysed reaction : :*Where S is substrate, P is product and C is catalyst. In the latter sense, it may refer to a surface on which other chemical reactions are performed or play a supporting role in a variety of spectroscopic and micro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tryptophan
Tryptophan (symbol Trp or W) is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, making it a polar molecule with a non-polar aromatic beta carbon substituent. Tryptophan is also a precursor to the neurotransmitter serotonin, the hormone melatonin, and vitamin B3 (niacin). It is encoded by the codon UGG. Like other amino acids, tryptophan is a zwitterion at physiological pH where the amino group is protonated (–; pKa = 9.39) and the carboxylic acid is deprotonated ( –COO−; pKa = 2.38). Humans and many animals cannot synthesize tryptophan: they need to obtain it through their diet, making it an essential amino acid. Tryptophan is named after the digestive enzymes trypsin, which were used in its first isolation from casein proteins. It was assigned the one-letter symbol W based on the double ring being visually suggestive to the bulky letter. Function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG). Proline is the only proteinogenic amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring. History and etymology Proline was first isolated in 1900 by Richard Willstätter who obtained the amino a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
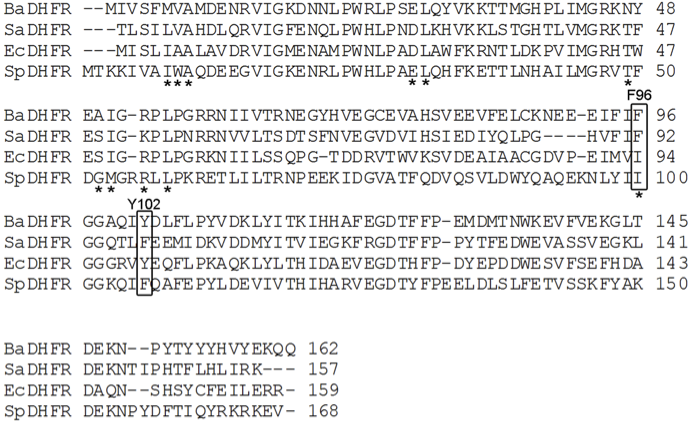
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst eukaryotes, and the tmRNA in bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 194 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that j ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding site'', and residues that catalyse a reaction of that substrate, the ''catalytic site''. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their functio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |