|
Mobile Genetic Elements
Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome are thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. These mechanisms can also rearrange genes in the host genome generating variation. These mechanisms can increase fitness by gaining new or additional functions. An example of MGEs in evolutionary context are that virulence factors and antibiotic resistance genes of MGEs can be transported to share genetic code with neighboring bacteria. However, MGEs can also decrease fitness by introducing disease-causing alleles or mutations. The set of MGEs in an organism is called a mobil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Components Of The Human Genome
Component may refer to: In engineering, science, and technology Generic systems *System components, an entity with discrete structure, such as an assembly or software module, within a system considered at a particular level of analysis * Lumped element model, a model of spatially distributed systems Electrical *Component video, a type of analog video information that is transmitted or stored as two or more separate signals *Electronic component, a constituent of an electronic circuit *Symmetrical components, in electrical engineering, analysis of unbalanced three-phase power systems Mathematics *Color model, a way of describing how colors can be represented, typically as multiple values or color components * Component (group theory), a quasi-simple subnormal sub-group *Connected component (graph theory), a maximal connected subgraph *Connected component (topology), a maximal connected subspace of a topological space *Vector component, result of the decomposition of a vector into ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome. Recombinant DNA is the general name for a piece of DNA that has been created by combining two or more fragments from different sources. Recombinant DNA is possible because DNA molecules from all organisms share the same chemical structure, differing only in the nucleotide sequence. Recombinant DNA molecules are sometimes called chimeric DNA because they can be made of material from two different species like the mythical chimera. rDNA technology uses palindromic sequences and leads to the production of sticky and blunt ends. The DNA sequences used in the construction of recombinant DNA molecules can originate from any species. For example, plant DNA can be joined to bacterial DNA, or human DNA can be joined with fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus (genetics)
In genetics, a locus (: loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of Human genome#Coding sequences (protein-coding genes), protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygote, homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygote, heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transposition. The word "transposase" was first coined by the individuals who cloned the enzyme required for transposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that some segments of chromosomes changed their position, jumping between different loci or from one chromosome to another. The repositioning of these transposons (which coded for color) allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Transposons
DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I TEs, retrotransposons, that move through an RNA intermediate. DNA transposons can move in the DNA of an organism via a single-or double-stranded DNA intermediate. DNA transposons have been found in both Prokaryote, prokaryotic and Eukaryote, eukaryotic organisms. They can make up a significant portion of an organism's genome, particularly in eukaryotes. In prokaryotes, TE's can facilitate the Horizontal gene transfer, horizontal transfer of antibiotic resistance or other genes associated with virulence. After replicating and propagating in a host, all transposon copies become inactivated and are lost unless the transposon passes to a genome by starting a new life cycle with horizontal transfer. DNA transposons do not randomly insert themse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Giant Viruses
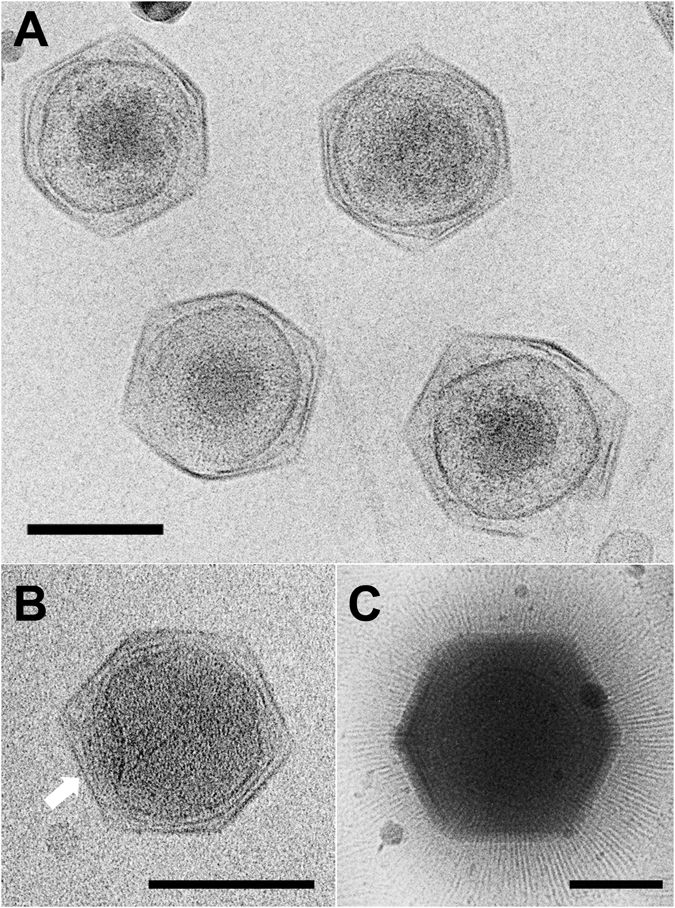
A giant virus, sometimes referred to as a girus, is a very large virus, some of which are larger than typical bacteria. All known giant viruses belong to the phylum '' Nucleocytoviricota''. Description While the exact criteria as defined in the scientific literature vary, giant viruses are generally described as viruses having large, pseudo- icosahedral capsids (200 to 400 nanometers in diameter) that may be surrounded by a thick (approximately 100 nm) layer of filamentous protein fibers. The viruses have large, double-stranded DNA genomes (300 to >1000 kilobasepairs) that encode a large contingent of genes (of the order of 1000 genes). The best characterized giant viruses are the phylogenetically related mimivirus and megavirus, which belong to the family ''Mimiviridae'' (aka ''Megaviridae''), and are distinguished by their large capsid diameters. Giant viruses from the deep ocean, terrestrial sources, and human patients contain genes encoding cytochrome P450 (CYP; P450) en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Organism
An organism is any life, living thing that functions as an individual. Such a definition raises more problems than it solves, not least because the concept of an individual is also difficult. Many criteria, few of them widely accepted, have been proposed to define what an organism is. Among the most common is that an organism has autonomous reproduction, Cell growth, growth, and metabolism. This would exclude viruses, despite the fact that they evolution, evolve like organisms. Other problematic cases include colonial organisms; a colony of eusocial insects is organised adaptively, and has Germ-Soma Differentiation, germ-soma specialisation, with some insects reproducing, others not, like cells in an animal's body. The body of a siphonophore, a jelly-like marine animal, is composed of organism-like zooids, but the whole structure looks and functions much like an animal such as a jellyfish, the parts collaborating to provide the functions of the colonial organism. The evolutiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Symbiosis
Symbiosis (Ancient Greek : living with, companionship < : together; and ''bíōsis'': living) is any type of a close and long-term biological interaction, between two organisms of different species. The two organisms, termed symbionts, can for example be in Mutualism (biology), mutualistic, commensalism, commensalistic, or parasitism, parasitic relationships. In 1879, Heinrich Anton de Bary defined symbiosis as "the living together of unlike organisms". The term is sometimes more exclusively used in a restricted, mutualistic sense, where both symbionts contribute to each other's subsistence. This means that they benefit each other in some way. Symbiosis can be ''obligate'' (or ''obligative''), which means that one, or both of the organisms depend on each other for survival, or ''facultative'' (optional), when they can also subsist independently. Symbiosis is also classified by physical attachment. Symbionts forming a single body live ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequences
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure. The sequence represents genetic information. Biological deoxyribonucleic acid represents the information which directs the functions of an organism. Nucleic acids also have a secondary structure and tertiary structure. Primary structure is sometimes mistakenly referred to a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposons
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome. The discovery of mobile genetic elements earned Barbara McClintock a Nobel Prize in 1983. There are at least two classes of TEs: Class I TEs or retrotransposons generally function via reverse transcription, while Class II TEs or DNA transposons encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. Discovery by Barbara McClintock Barbara McClintock discovered the first TEs in maize (''Zea mays'') at the Cold Spring Harbor Laboratory in New York. McClintock was experimenting with maize plants that had broken chromosomes. In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (Style (botany), style) of the flower received pollen from its own anther. These kernels came fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cut And Paste Mechanism Of Transposition
Cut or CUT may refer to: Common uses * The act of cutting, the separation of an object into two through acutely directed force ** A type of wound ** Cut (archaeology), a hole dug in the past ** Cut (clothing), the style or shape of a garment ** Cut (earthworks), an excavation to make way for a transport route ** Cut (gems) ** Cut of meat * Cut (etiquette), a snub or slight such as failure to greet an acquaintance Geography Romania * Cut, Alba, a commune * Cut, a village in Dumbrava Roșie United States * Cut, Texas, an unincorporated community * Cut River (Mackinac County, Michigan) * Cut River (Roscommon County, Michigan) * Custer County Airport, South Dakota, US (FAA identifier CUT) Computing and mathematics * Cut (logic programming) * cut (Unix), a command line utility * Cut, copy, and paste, a set of editing procedures * Control Unit Terminal, a kind of IBM display terminal for mainframe computers * Cut (graph theory) Books * ''Cut'' (manga), a 2008 Japanese ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |