|
Chromodomain Helicase DNA-binding (CHD) Protein Subfamily
Chromodomain helicase DNA-binding (CHD) proteins is a subfamily of ATP-dependent chromatin remodeling complexes (remodelers). All remodelers fall under the umbrella of RNA/DNA helicase superfamily 2. In yeast, CHD complexes are primarily responsible for nucleosome assembly and organization. These complexes play an additional role in multicellular eukaryotes, assisting in chromatin access and nucleosome editing. Functions of CHD subfamily proteins Similar to the imitation switch (ISWI) subfamily of ATP-dependent chromatin remodelers, CHD complexes regulate the assembly and organization of mature nucleosomes along the DNA. Histones are removed during DNA replication; following behind the replisome, histones start to assemble as immature pre-nucleosomes on nascent DNA. With the help of CHD complexes, histone octamers can mature into native nucleosomes. Following nucleosome formation, CHD complexes organize nucleosomes by regularly spacing them apart along the DNA. Additionally, CHDs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATP-dependent Chromatin Remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
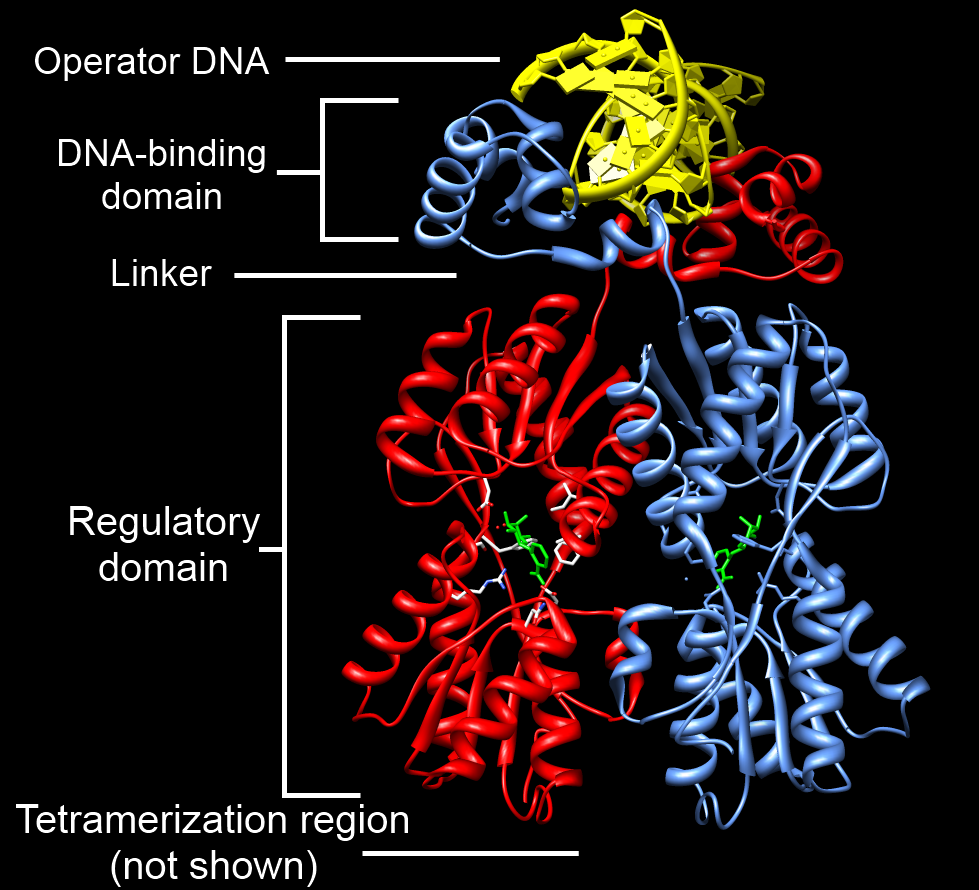
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD7
Chromodomain-helicase-DNA-binding protein 7 also known as ATP-dependent helicase CHD7 is an enzyme that in humans is encoded by the ''CHD7'' gene. CHD7 is an ATP-dependent chromatin remodeler homologous to the Drosophila trithorax-group protein Kismet. Mutations in CHD7 are associated with CHARGE syndrome. This protein belongs to a larger group of ATP-dependent chromatin remodeling complexes, the CHD subfamily. Model organisms Model organisms have been used in the study of CHD7 function. A conditional knockout mouse line, called ''Chd7tm2a(EUCOMM)Wtsi'' was generated as part of the International Knockout Mouse Consortium program — a high-throughput mutagenesis project to generate and distribute animal models of disease to interested scientists. Male and female animals underwent a standardized phenotypic screen to determine the effects of deletion. Twenty four tests were carried out on mutant mice and five significant abnormalities were observed. No homozygous mutant em ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD5
Chromodomain-helicase-DNA-binding protein 5 is an enzyme that in humans is encoded by the ''CHD5'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... It is a part of the CHD subfamily of ATP-dependent chromatin remodeling complexes. References External links * Further reading * * * * * * * * * {{gene-1-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD3
Chromodomain-helicase-DNA-binding protein 3 is an enzyme that in humans is encoded by the ''CHD3'' gene. Function This gene encodes a member of the CHD family of proteins which are characterized by the presence of chromo (chromatin organization modifier) domains and SNF2-related helicase/ATPase domains. This protein is one of the components of a histone deacetylase complex referred to as the Mi-2/NuRD complex which participates in the remodeling of chromatin by deacetylating histones. Chromatin remodeling is essential for many processes including transcription. Autoantibodies against this protein are found in a subset of patients with dermatomyositis. Three alternatively spliced transcripts encoding different isoforms have been described. Mutations in CHD3 cause a neurodevelopmental syndrome ( Snijders Blok-Campeau syndrome) with macrocephaly and impaired speech and language. Interactions CHD3 has been shown to interact with: * HDAC1, * Histone deacetylase 2 Histone deac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD2
Chromodomain-helicase-DNA-binding protein 2 is an enzyme that in humans is encoded by the ''CHD2'' gene. Function The CHD family of proteins is characterized by the presence of chromo (chromatin organization modifier) domains and SNF2-related helicase/ATPase domains. CHD genes alter gene expression possibly by modification of chromatin structure thus altering access of the transcriptional apparatus to its chromosomal DNA template. CHD2 catalyzes the assembly of chromatin into periodic arrays; and the N-terminal region of CHD2, which contains tandem chromodomains, serves an auto-inhibitory role in both the DNA-binding and ATPase activities of CHD2. Alternatively spliced transcript variants encoding distinct isoforms A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some iso ... have been ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD1
The Chromodomain-Helicase DNA-binding 1 is a protein that, in humans, is encoded by the CHD1 gene. CHD1 is a chromatin remodeling protein that is widely conserved across many eukaryotic organisms, from yeast to humans. CHD1 is named for three of its protein domains: two tandem chromodomains, its ATPase catalytic domain, and its DNA-binding domain (Figure 1). The CHD1 remodeler binds nucleosomes and induces local changes in nucleosome positioning through ATP hydrolysis coupled to DNA translocation of the DNA across the histone proteins. The catalytic domain of CHD1, which is highly conserved across all nucleosome remodelers, is a two-lobed structure. CHD1 relies on the DNA-binding domain, which binds DNA in a sequence non-specific manner, to help regulate spacing. CHD1 is a member of a large family of CHD nucleosome remodelers, though yeast has only one CHD protein, called Chd1. Humans and mice, by contrast, have ten CHD proteins that are homologous to CHD1, but each have their own ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RecA
RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea. RecA has multiple activities, all related to DNA repair. In the bacterial SOS response, it has a co-protease function in the autocatalytic cleavage of the LexA repressor and the λ repressor. RecA's association with DNA repair is based on its central role in homologous recombination. The RecA protein binds strongly and in long clusters to ssDNA to form a nucleoprotein filament. The protein has more than one DNA binding site, and thus can hold a single strand and double strand together. This feature makes it possible to catalyze a DNA synapsis reaction between a DNA double helix and a complementary region of single-stranded DNA. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromodomain
A chromodomain (''chromatin organization modifier'') is a protein structural domain of about 40–50 amino acid residues commonly found in proteins associated with the remodeling and manipulation of chromatin. The domain is highly conserved among both plants and animals, and is represented in a large number of different proteins in many genomes, such as that of the mouse. Some chromodomain-containing genes have multiple alternative splicing isoforms that omit the chromodomain entirely. In mammals, chromodomain-containing proteins are responsible for aspects of gene regulation related to chromatin remodeling and formation of heterochromatin regions. Chromodomain-containing proteins also bind methylated histones and appear in the RNA-induced transcriptional silencing complex. In histone modifications, chromodomains are very conserved. They function by identifying and binding to methylated lysine residues that exist on the surface of chromatin proteins and thereby regulate g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Functional Domains Of The CHD Subfamily Of ATP-dependent Chromatin Remodellers
Functional may refer to: * Movements in architecture: ** Functionalism (architecture) ** Form follows function * Functional group, combination of atoms within molecules * Medical conditions without currently visible organic basis: ** Functional symptom ** Functional disorder * Functional classification for roads * Functional organization * Functional training In mathematics * Functional (mathematics), a term applied to certain scalar-valued functions in mathematics and computer science ** Functional analysis ** Linear functional, a type of functional often simply called a functional in the context of functional analysis * Higher-order function, also called a functional, a function that takes other functions as arguments In computer science, software engineering * (C++), a header file in the C++ Standard Library * Functional design, a paradigm used to simplify the design of hardware and software devices * Functional model, a structured representation of functions, activit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expressi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |