|
Cajal Bodies
Cajal bodies (CBs), also coiled bodies, are spherical nuclear bodies of 0.3–1.0 μm in diameter found in the nucleus of proliferative cells like embryonic cells and tumor cells, or metabolically active cells like neurons. CBs are membrane-less organelles and largely consist of proteins and RNA. They were first reported by Santiago Ramón y Cajal in 1903, who called them ''nucleolar accessory bodies'' due to their association with the nucleoli in neuronal cells. They were rediscovered with the use of the electron microscope (EM) and named ''coiled bodies'', according to their appearance as coiled threads on EM images, and later renamed after their discoverer. Research on CBs was accelerated after discovery and cloning of the marker protein p80/Coilin. CBs have been implicated in RNA-related metabolic processes such as the biogenesis, maturation and recycling of snRNPs, histone mRNA processing and telomere maintenance. CBs assemble RNA which is used by telomerase to add nucle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly ''Drosophila melanogaster'' lacks telomerase, but instead uses retrotransposons to maintain telomeres. Telomerase is a reverse transcriptase enzyme that carries its own RNA molecule (e.g., with the sequence 3′- CCC AA UCCC-5′ in '' Trypanosoma brucei'') which is used as a template when it elongates telomeres. Telomerase is active in gametes and most cancer cells, but is normally absent in most somatic cells. History The existence of a compensatory mechanism for telomere shortening was first found by Soviet biologist Alexey Olovnikov in 1973, who also suggested the telomere hypothesis of aging and the telo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear genes, nuclear-encoded genes, splicing occurs in the cell nucleus, nucleus either during or immediately after Transcription (biology), transcription. For those eukaryotic transcription, eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translation (biology), translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Component Of Telomerase
Telomerase RNA component, also known as TR, TER or TERC, is an ncRNA found in eukaryotes that is a component of telomerase, the enzyme used to extend telomeres. TERC serves as a template for telomere replication (reverse transcription) by telomerase. Telomerase RNAs differ greatly in sequence and structure between vertebrates, ciliates and yeasts, but they share a 5' pseudoknot structure close to the template sequence. The vertebrate telomerase RNAs have a 3' H/ACA snoRNA-like domain. Structure TERC is a Long non-coding RNA (lncRNA) ranging in length from ~150nt in ciliates to 400-600nt in vertebrates, and 1,300nt in yeast (Alnafakh). Mature human TERC (hTR) is 451nt in length. TERC has extensive secondary structural features over 4 principal conserved domains. The core domain, the largest domain at the 5’ end of TERC, contains the CUAAC Telomere template sequence. Its secondary structure consists of a large loop containing the template sequence, a P1 loop-closing heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ScaRNA
Small Cajal body-specific RNAs (scaRNAs) are a class of small nucleolar RNAs (snoRNAs) that specifically localise to the Cajal body, a nuclear organelle (cellular sub-organelle) involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs or snurps). ScaRNAs guide the modification (methylation and pseudouridylation) of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12. The first scaRNA identified was U85. It is unlike typical snoRNAs in that it is a composite C/D box and H/ACA box snoRNAs and can guide both pseudouridylation and 2′-''O''-methylation. Not all scaRNAs are composite C/D and H/ACA box snoRNA and most scaRNAs are structurally and functionally indistinguishable from snoRNAs, directing ribosomal RNA (rRNA) modification in the nucleolus. ''Drosophila'' scaRNAs Several studies identified scaRNAs in ''Drosophila.'' One of the studies showed that several ''Drosophila'' scaRNAs could function equally well in the nucleoplasm of mut ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coilin
Coilin is a protein that in humans is encoded by the ''COIL'' gene. Coilin got its name from the coiled shape of the Cajal bodies in which it is found. It was first identified using human autoimmune serum. Function Coilin protein is one of the main molecular components of Cajal bodies. Cajal bodies are non-membrane bound nuclear bodies of varying number and composition that are involved in the post-transcriptional modification of small nuclear and small nucleolar RNAs. In addition to its structural role, coilin acts as glue to connect the CB to the nucleolus. The N-terminus of the coilin protein directs its self-oligomerization while the C-terminus influences the number of nuclear bodies assembled per cell. Differential methylation and phosphorylation of coilin likely influences its localization among nuclear bodies and the composition and assembly of Cajal bodies. This gene has pseudogenes on chromosome 4 and chromosome 14. To study CBs, coilin can be combined with GFP (Gree ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolus
The nucleolus (; : nucleoli ) is the largest structure in the cell nucleus, nucleus of eukaryote, eukaryotic cell (biology), cells. It is best known as the site of ribosome biogenesis. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of the nucleolus is the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Theodor Schwann in his 1839 treatise described that Matthias Jakob Schleiden, Schleiden had identified small corpuscles in nuclei, and named the structures "Kernkörperchen". In a 1947 translation of the work to English, the structure was named "nucleolus". Little was known about the fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many RNA viruses is composed of negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process is catalyzed by a viral RNA dependent RNA polymerase. Background A DNA transcription unit encoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
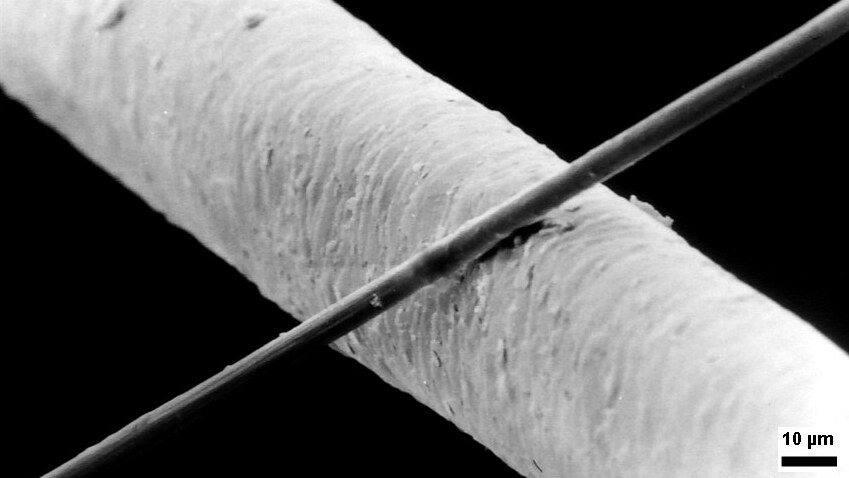
Micrometre
The micrometre (English in the Commonwealth of Nations, Commonwealth English as used by the International Bureau of Weights and Measures; SI symbol: μm) or micrometer (American English), also commonly known by the non-SI term micron, is a unit of length in the International System of Units (SI) equalling (SI standard prefix "micro-" = ); that is, one millionth of a metre (or one thousandth of a millimetre, , or about ). The nearest smaller common SI Unit, SI unit is the nanometre, equivalent to one thousandth of a micrometre, one millionth of a millimetre or one billionth of a metre (). The micrometre is a common unit of measurement for wavelengths of infrared radiation as well as sizes of biological cell (biology), cells and bacteria, and for grading wool by the diameter of the fibres. The width of a single human hair ranges from approximately 20 to . Examples Between 1 μm and 10 μm: * 1–10 μm – length of a typical bacterium * 3–8 μm – width of str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalysis, catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks Covalent bond, bonds. Proteases are involved in numerous biological pathways, including Digestion#Protein digestion, digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently convergent evolution, evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Classification Based on catalytic residue Proteases can be classified into seven broad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
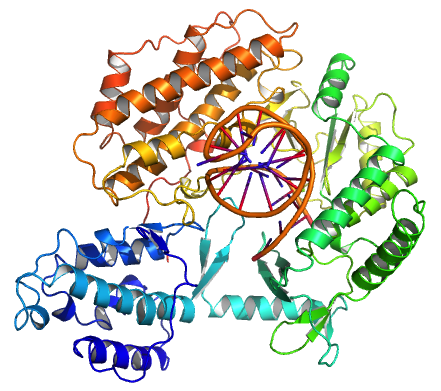
Ribonucleoproteins
Nucleoproteins are proteins conjugated with nucleic acids (either DNA or RNA). Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins. Structures Nucleoproteins tend to be positively charged, facilitating interaction with the negatively charged nucleic acid chains. The tertiary structures and biological functions of many nucleoproteins are understood.Graeme K. Hunter G. K. (2000): Vital Forces. The discovery of the molecular basis of life. Academic Press, London 2000, . Important techniques for determining the structures of nucleoproteins include X-ray diffraction, nuclear magnetic resonance and cryo-electron microscopy. Viruses Virus genomes (either DNA or RNA) are extremely tightly packed into the viral capsid. Many viruses are therefore little more than an organised collection of nucleoproteins with their binding sites pointing inwards. Structurally characterised viral nucleoproteins include influenza, rabies, Ebola, Bunyamwera, Schmallenb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |