|
Branched-chain Alpha-keto Acid Dehydrogenase Complex
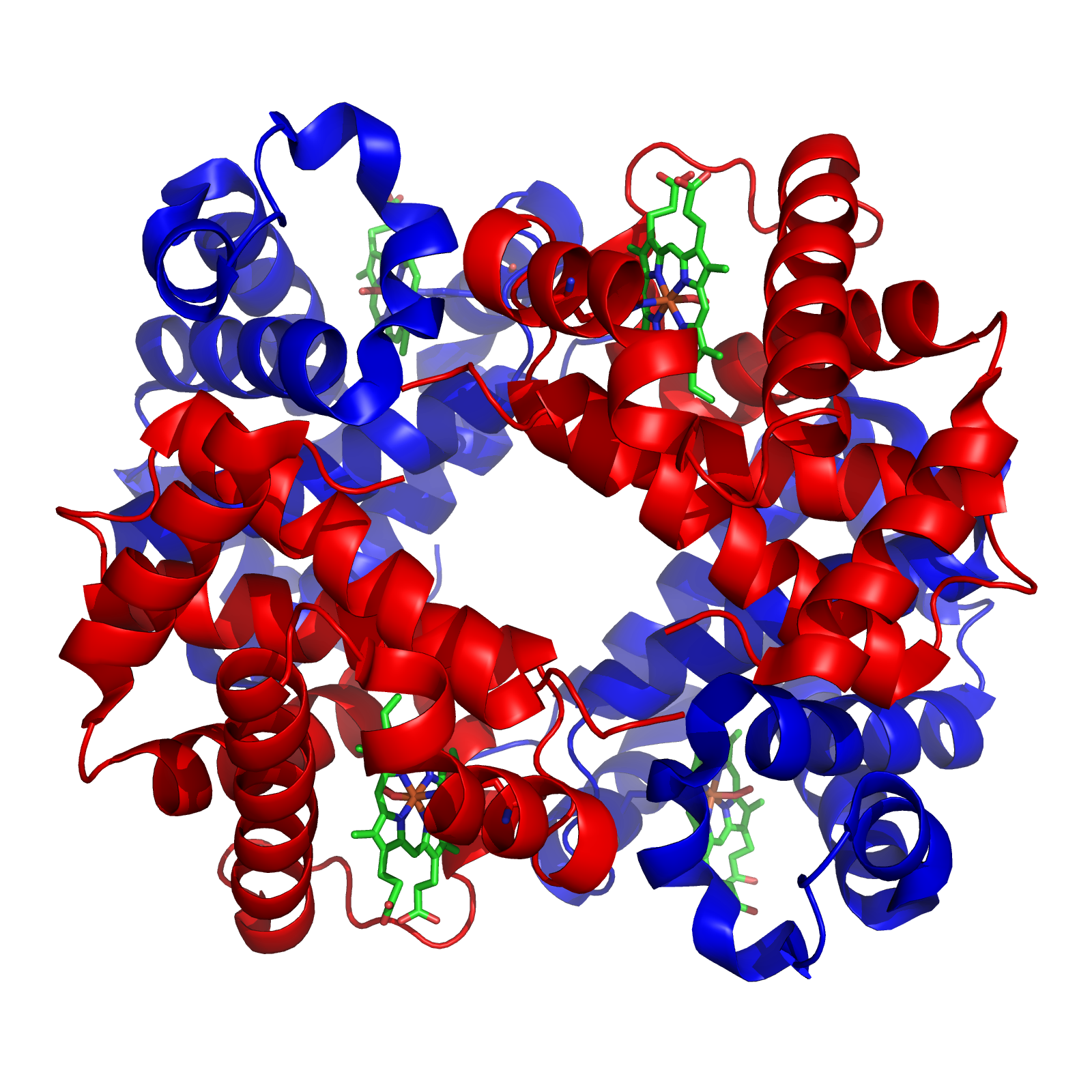
The branched-chain α-ketoacid dehydrogenase complex (BCKDC or BCKDH complex) is a multi-subunit complex of enzymes that is found on the mitochondrial inner membrane. This enzyme complex catalyzes the oxidative decarboxylation of branched, short-chain alpha-ketoacids. BCKDC is a member of the mitochondrial α-ketoacid dehydrogenase complex family, which also includes pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase, key enzymes that function in the Krebs cycle. Biological function In animal tissue, BCKDC catalyzes an irreversible step in the catabolism of the branched-chain amino acids L-isoleucine, L-valine, and L-leucine, acting on their deaminated derivatives (L-alpha-keto-beta-methylvalerate, alpha-ketoisovalerate, and alpha-ketoisocaproate, respectively) and converting them to α-Methylbutyryl-CoA, Isobutyryl-CoA and Isovaleryl-CoA respectively. In bacteria, this enzyme participates in the synthesis of branched, long-chain fatty acids. In plants, this enzyme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thiamine Pyrophosphate
Thiamine pyrophosphate (TPP or ThPP), or thiamine diphosphate (ThDP), or cocarboxylase is a thiamine (vitamin B1) derivative which is produced by the enzyme thiamine diphosphokinase. Thiamine pyrophosphate is a cofactor that is present in all living systems, in which it catalyzes several biochemical reactions. Thiamine pyrophosphate is synthesized in the cytosol and is required in the cytosol for the activity of transketolase and in the mitochondria for the activity of pyruvate-, oxoglutarate- and branched chain keto acid dehydrogenases. To date, the yeast ThPP carrier (Tpc1p) the human Tpc and the ''Drosophila melanogaster'' have been identified as being responsible for the mitochondrial transport of ThPP and ThMP. It was first discovered as an essential nutrient (vitamin) in humans through its link with the peripheral nervous system disease beriberi, which results from a deficiency of thiamine in the diet. TPP works as a coenzyme in many enzymatic reactions, such as: * Pyru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding site'', and residues that catalyse a reaction of that substrate, the ''catalytic site''. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their functio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acyltransferase
Acyltransferase is a type of transferase enzyme that acts upon acyl In chemistry, an acyl group is a moiety derived by the removal of one or more hydroxyl groups from an oxoacid, including inorganic acids. It contains a double-bonded oxygen atom and an organyl group () or hydrogen in the case of formyl grou ... groups. Examples include: * Glycerol-3-phosphate acyltransferases * Glyceronephosphate O-acyltransferase * Lecithin-cholesterol acyltransferase * Long-chain-alcohol O-fatty-acyltransferase See also * Acetyltransferase External links * Transferases EC 2.3 {{2.3-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carboxy-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that j ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homodimers
In biochemistry, a protein dimer is a macromolecular complex or multimer formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins while a protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetramers
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula Ti(OCH3)4, which is tetrameric in solid state and has the molecular formula Ti4(OCH3)16. An example from organic chemistry is kobophenol A, a substance that is formed by combining four molecules of resveratrol. In biochemistry, it similarly refers to a biomolecule formed of four units, that are the same ( homotetramer), i.e. as in Concanavalin A or different ( heterotetramer), i.e. as in hemoglobin. Hemoglobin has 4 similar sub-units while immunoglobulins have 2 very different sub-units. The different sub-units may have each their own activity, such as binding biotin in avidin tetramers, or have a common biological property, such as the allosteric binding of oxygen Oxygen is a chemical element; it has chemical symbol, symbol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymer
A polymer () is a chemical substance, substance or material that consists of very large molecules, or macromolecules, that are constituted by many repeat unit, repeating subunits derived from one or more species of monomers. Due to their broad spectrum of properties, both synthetic and natural polymers play essential and ubiquitous roles in everyday life. Polymers range from familiar synthetic plastics such as polystyrene to natural biopolymers such as DNA and proteins that are fundamental to biological structure and function. Polymers, both natural and synthetic, are created via polymerization of many small molecules, known as monomers. Their consequently large molecular mass, relative to small molecule compound (chemistry), compounds, produces unique physical property, physical properties including toughness, high rubber elasticity, elasticity, viscoelasticity, and a tendency to form Amorphous solid, amorphous and crystallization of polymers, semicrystalline structures rath ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nicotinamide Adenine Dinucleotide
Nicotinamide adenine dinucleotide (NAD) is a Cofactor (biochemistry), coenzyme central to metabolism. Found in all living cell (biology), cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an Redox, oxidized and reduced form, abbreviated as NAD and NADH (H for hydrogen), respectively. In cellular metabolism, NAD is involved in redox reactions, carrying electrons from one reaction to another, so it is found in two forms: NAD is an oxidizing agent, accepting electrons from other molecules and becoming reduced; with H+, this reaction forms NADH, which can be used as a reducing agent to donate electrons. These electron transfer reactions are the main function of NAD. It is also used in other cellular processes, most notably as a substrate (biochemistry), substrate of enzymes in adding or removing chemical groups to or fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flavin Adenine Dinucleotide
In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex. FAD can exist in four redox states, which are the flavin-N(5)-oxide, quinone, semiquinone, and hydroquinone. FAD is converted between these states by accepting or donating electrons. FAD, in its fully oxidized form, or quinone form, accepts two electrons and two protons to become FADH2 (hydroquinone form). The semiquinone (FADH·) can be formed by either reduction of FAD or oxidation of FADH2 by accepting or donating one electron and one proton, respectively. Some proteins, however, generate and maintain a super ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrolipoamide Dehydrogenase
Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the ''DLD'' gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide. Dihydrolipoamide dehydrogenase (DLD) is a mitochondrial enzyme that plays a vital role in energy metabolism in eukaryotes. This enzyme is required for the complete reaction of at least five different multi-enzyme complexes. Additionally, DLD is a flavoenzyme oxidoreductase that contains a reactive disulfide bridge and a FAD cofactor that are directly involved in catalysis. The enzyme associates into tightly bound homodimers required for its enzymatic activity. File:Lipoamide-2D-skeletal.png, Lipoamide File:Dihydrolipoamide.svg, Dihydrolipoamide Structure The protein encoded by the DLD gene comes together with another protein to form a dimer in the central metabolic pathway. Several amino acids within the catalytic pocket have been identified ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |