|
Alcanivorax Pacificus
''Alcanivorax pacificus'' is a pyrene-degrading marine gammaproteobacterium. It is of the genus ''Alcanivorax'', a group of marine bacteria known for degrading hydrocarbons. When originally proposed, the genus ''Alcanivorax'' comprised six distinguishable species. However, ''A. pacificus'', a seventh strain, was isolated from deep sea sediments in the West Pacific Ocean by Shanghai Majorbio Bio-pharm Technology Co., Ltd. in 2011. ''A. pacificus’s'' ability to degrade hydrocarbons can be employed for cleaning up oil-contaminated oceans through bioremediation. The genomic differences present in this strain of ''Alcanivorax'' that distinguish it from the original consortium are important to understand to better utilize this bacteria for bioremediation. Characteristics ''A. pacificus'' is a non-motile, Gram-negative bacillus. It is oxidase- and catalase-positive. It forms light grey colonies that are between 1 and 2 millimeters in diameter when grown on 216L marine agar medium con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrene
Pyrene is a polycyclic aromatic hydrocarbon (PAH) consisting of four fused benzene rings, resulting in a flat aromatic system. The chemical formula is . This yellow solid is the smallest peri-fused PAH (one where the rings are fused through more than one face). Pyrene forms during incomplete combustion of organic compounds. Occurrence and properties Pyrene was first isolated from coal tar, where it occurs up to 2% by weight. As a peri-fused PAH, pyrene is much more resonance-stabilized than its five-member-ring containing isomer fluoranthene. Therefore, it is produced in a wide range of combustion conditions. For example, automobiles produce about 1 μg/km.Senkan, Selim and Castaldi, Marco (2003) "Combustion" in ''Ullmann's Encyclopedia of Industrial Chemistry'', Wiley-VCH, Weinheim. Reactions Oxidation with chromate affords perinaphthenone and then naphthalene-1,4,5,8-tetracarboxylic acid. Pyrene undergoes a series of hydrogenation reactions and is susceptible to halog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium Citrate
Sodium citrate may refer to any of the sodium salts of citric acid (though most commonly the third): * Monosodium citrate * Disodium citrate * Trisodium citrate The three forms of salt are collectively known by the E number E331. Applications Food Sodium citrates are used as acidity regulators in food and drinks, and also as emulsifiers for oils. They enable cheeses to melt without becoming greasy. It reduces the acidity of food as well. Blood clotting inhibitor Sodium citrate is used to prevent donated blood from clotting in storage. It is also used in a laboratory, before an operation, to determine whether a person's blood is too thick and might cause a blood clot, or if the blood is too thin to safely operate. Sodium citrate is used in medical contexts as an alkalinizing agent in place of sodium bicarbonate, to neutralize excess acid in the blood and urine. Metabolic acidosis It has applications for the treatment of metabolic acidosis and chronic kidney disease. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alkylation
Alkylation is the transfer of an alkyl group from one molecule to another. The alkyl group may be transferred as an alkyl carbocation, a free radical, a carbanion, or a carbene (or their equivalents). Alkylating agents are reagents for effecting alkylation. Alkyl groups can also be removed in a process known as dealkylation. Alkylating agents are often classified according to their nucleophilic or electrophilic character. In oil refining contexts, alkylation refers to a particular alkylation of isobutane with olefins. For upgrading of petroleum, alkylation produces a premium blending stock for gasoline. In medicine, alkylation of DNA is used in chemotherapy to damage the DNA of cancer cells. Alkylation is accomplished with the class of drugs called alkylating antineoplastic agents. Nucleophilic alkylating agents Nucleophilic alkylating agents deliver the equivalent of an alkyl anion ( carbanion). The formal "alkyl anion" attacks an electrophile, forming a new cova ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alkane Hydroxylase
In organic chemistry, an alkane, or paraffin (a historical trivial name that also has other meanings), is an acyclic saturated hydrocarbon. In other words, an alkane consists of hydrogen and carbon atoms arranged in a tree structure in which all the carbon–carbon bonds are single. Alkanes have the general chemical formula . The alkanes range in complexity from the simplest case of methane (), where ''n'' = 1 (sometimes called the parent molecule), to arbitrarily large and complex molecules, like pentacontane () or 6-ethyl-2-methyl-5-(1-methylethyl) octane, an isomer of tetradecane (). The International Union of Pure and Applied Chemistry (IUPAC) defines alkanes as "acyclic branched or unbranched hydrocarbons having the general formula , and therefore consisting entirely of hydrogen atoms and saturated carbon atoms". However, some sources use the term to denote ''any'' saturated hydrocarbon, including those that are either monocyclic (i.e. the cycloalkane ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alcanivorax Borkumensis
''Alcanivorax borkumensis'' is an alkane-degrading marine bacterium which naturally propagates and becomes predominant in crude-oil-containing seawater when nitrogen and phosphorus nutrients are supplemented. Description ''A. borkumensis'' is a rod-shaped bacterium without flagella that obtains its energy primarily from consuming alkanes (a type of hydrocarbon). It is aerobic, meaning it uses oxygen to gain energy, and it is halophilic, meaning it tends to live in environments that contain salt, such as salty ocean water. It is also Gram-negative, which essentially means it has a relatively thin cell wall. It is also nonmotile; however, other organisms that appear to be in the same genus are motile through flagella. Discovery The microorganism was discovered near the island of Borkum (hence the epithet ''borkumensis'') by the Helmholtz Centre for Infection Research and the Technical University of Braunschweig and in 2006, them and the University of Bielefeld identified the Bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alcanivorax Dieselolei
''Alcanivorax dieselolei'' is a species of alkane-degrading bacteria. Its genome has been sequenced. It is halophilic, aerobic, Gram-negative, non-spore-forming, catalase- and oxidase In biochemistry, an oxidase is an enzyme that catalyzes oxidation-reduction reactions, especially one involving dioxygen (O2) as the electron acceptor. In reactions involving donation of a hydrogen atom, oxygen is reduced to water (H2O) or hydro ...-positive, motile and rod-shaped. Its type strain is B-5T (=DSM 16502T =CGMCC 1.3690T). References Further reading * External links *LPSN [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S RRNA
16S rRNA may refer to: * 16S ribosomal RNA 16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S r ..., the prokaryotic ribosomal subunit * Mitochondrially encoded 16S RNA, the eukaryotic ribosomal subunit {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but since the discovery of the first operons in eukaryotes in the early 1990s, more evidence has arisen to suggest they are more common than previously assumed. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthologous
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
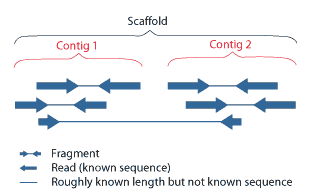
Contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GC-content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of an implied four total bases, also including adenine and thymine in DNA and adenine and uracil in RNA. GC-content may be given for a certain fragment of DNA or RNA or for an entire genome. When it refers to a fragment, it may denote the GC-content of an individual gene or section of a gene (domain), a group of genes or gene clusters, a non-coding region, or a synthetic oligonucleotide such as a primer. Structure Qualitatively, guanine (G) and cytosine (C) undergo a specific hydrogen bonding with each other, whereas adenine (A) bonds specifically with thymine (T) in DNA and with uracil (U) in RNA. Quantitatively, each GC base pair is held together by three hydrogen bonds, while AT and AU base pairs are held together by two hydrogen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solexa
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets. Illumina's technology had purportedly reduced the cost of sequencing a human genome to by 2014. Its customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumina ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)