|
Adaptive Immunity In Jawless Vertebrates
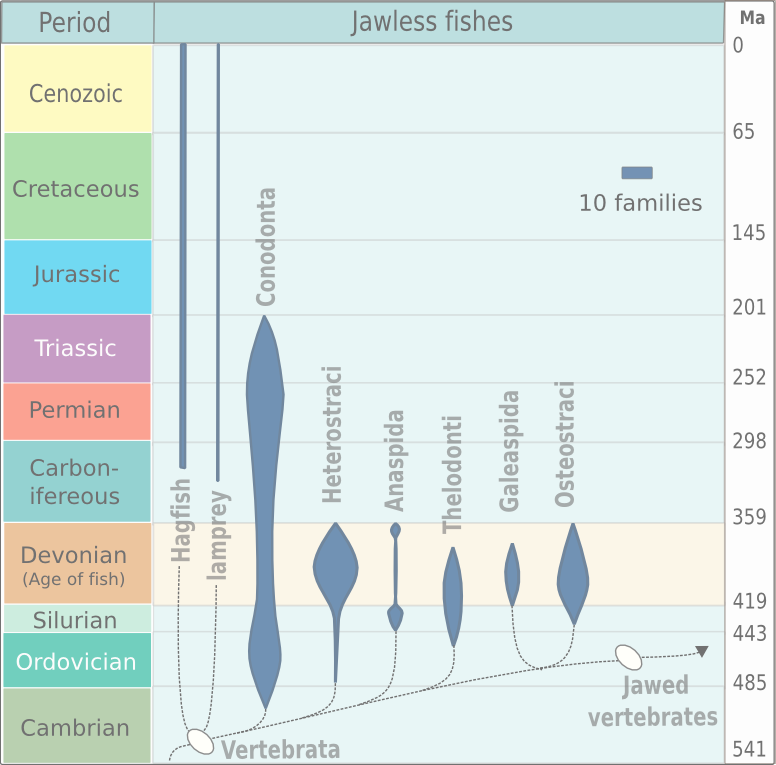
Jawless vertebrates, which today consist entirely of lampreys and hagfish, have an adaptive immune system similar to that found in jawed vertebrates. The cells of the agnathan AIS have roles roughly equivalent to those of B-cells and T-cells, with three lymphocyte lineages identified so far: *VLRA (most similar to α/β T cells, in its role and pathway of differentiation) *VLRB (most similar to B cells) *VLRC (most similar to γ/δ T cells) VLRA and VLRB were identified in 2009, while VLRC was discovered in 2013. Instead of immunoglobulins, they use variable lymphocyte receptors. Antigen receptors Jawless vertebrates do not have immunoglobulins (Igs), the key proteins to B-cells and T-cells. However, they do possess a system of leucine-rich repeat (LRR) proteins that make up variable lymphocyte receptors (VLRs). This system can produce roughly the same number of potential receptors that the Ig-based system found in jawed vertebrates can. Instead of recombination-activating gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Agnatha
Agnatha (; ) or jawless fish is a paraphyletic infraphylum of animals in the subphylum Vertebrata of the phylum Chordata, characterized by the lack of jaws. The group consists of both extant taxon, living (Cyclostomi, cyclostomes such as hagfishes and lampreys) and Extinction, extinct clades (e.g. conodonts and Cephalaspidomorphi, cephalaspidomorphs, among others). They are sister taxon, sister to vertebrates with jaws known as gnathostomes, who evolution, evolved from jawless ancestors during the early Silurian by developing folding joint, articulations in the first pairs of gill arches. Sequencing, Molecular data, both from rRNA and from mtDNA as well as Embryology, embryological data, strongly supports the hypothesis that both groups of living agnathans, hagfishes and lampreys, are more closely related to each other than to Gnathostomata, jawed fish, forming the Class (biology), superclass Cyclostomi. The oldest fossil agnathans appeared in the Cambrian. Living jawless fish c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombination-activating Gene
The recombination-activating genes (RAGs) encode parts of a protein complex that plays important roles in the rearrangement and recombination of the genes encoding immunoglobulin and T cell receptor molecules. There are two recombination-activating genes RAG1 and RAG2, whose cellular expression is restricted to lymphocytes during their developmental stages. The enzymes encoded by these genes, RAG-1 and RAG-2, are essential to the generation of mature B cells and T cells, two types of lymphocyte that are crucial components of the adaptive immune system. Function In the vertebrate immune system, each antibody is customized to attack one particular antigen (foreign proteins and carbohydrates) without attacking the body itself. The human genome has at most 30,000 genes, and yet it generates millions of different antibodies, which allows it to be able to respond to invasion from millions of different antigens. The immune system generates this diversity of antibodies by shuffling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
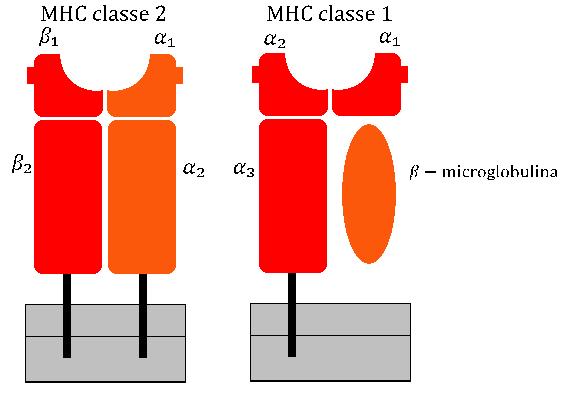
Major Histocompatibility Complex
The major histocompatibility complex (MHC) is a large Locus (genetics), locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for Cell (biology), cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules. Its name comes from its discovery during the study of transplanted tissue compatibility. Later studies revealed that tissue rejection due to incompatibility is only a facet of the full function of MHC molecules, which is to bind an antigen derived from self-proteins, or from pathogens, and bring the antigen presentation to the cell surface for recognition by the appropriate T cell, T-cells. MHC molecules mediate the interactions of leukocytes, also called white blood cells (WBCs), with other leukocytes or with body cells. The MHC determines donor compatibility for organ transplant, as well as one's susceptibility to autoimmune diseases. In a cell, protein molecules of the host's own pheno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T-cell Receptor
The T-cell receptor (TCR) is a protein complex, located on the surface of T cells (also called T lymphocytes). They are responsible for recognizing fragments of antigen as peptides bound to major histocompatibility complex (MHC) molecules. The binding between TCR and antigen peptides is of relatively low affinity and is biologically degenerate (that is, many TCRs recognize the same antigen peptide, and many antigen peptides are recognized by the same TCR). The TCR is composed of two different protein chains (that is, it is a hetero dimer). In humans, in 95% of T cells the TCR consists of an alpha (α) chain and a beta (β) chain (encoded by '' TRA'' and ''TRB'', respectively), whereas in 5% of T cells the TCR consists of gamma and delta (γ/δ) chains (encoded by '' TRG'' and '' TRD'', respectively). This ratio changes during ontogeny and in diseased states (such as leukemia). It also differs between species. Orthologues of the 4 loci have been mapped in various speci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptococcus Pneumoniae
''Streptococcus pneumoniae'', or pneumococcus, is a Gram-positive, spherical bacteria, hemolysis (microbiology), alpha-hemolytic member of the genus ''Streptococcus''. ''S. pneumoniae'' cells are usually found in pairs (diplococci) and do not form Bacterial morphological plasticity, spores and are non motile. As a significant human pathogenic bacterium ''S. pneumoniae'' was recognized as a major cause of pneumonia in the late 19th century, and is the subject of many humoral immunity studies. ''Streptococcus pneumoniae'' resides asymptomatically in healthy carriers typically colonizing the respiratory tract, sinuses, and nasopharynx, nasal cavity. However, in susceptible individuals with immunocompromised, weaker immune systems, such as the elderly and young children, the bacterium may become pathogenic and spread to other locations to cause disease. It spreads by direct person-to-person contact via respiratory droplets and by auto inoculation in persons carrying the bacteria in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Salmonella Typhimurium
''Salmonella enterica'' subsp. ''enterica'' is a subspecies of ''Salmonella enterica'', the rod-shaped, flagellated, aerobic, Gram-negative bacterium. Many of the pathogenic serovars of the ''S. enterica'' species are in this subspecies, including that responsible for typhoid. Serovars ''Salmonella enterica'' subsp. ''enterica'' serovars are defined based on their somatic (O) and flagellar (H) antigens, with over 2,600 serovars in total; only about 50 of these serovars are common causes of infections in humans. Most of these serovars are found in the environment and survive in plants, water, and soil; many serovars have broad host ranges that allow them to colonize different species in mammals, birds, reptiles, amphibians, and insects. Zoonotic diseases, like ''Salmonella'', spread between the environment and people. A number of techniques are currently used to differentiate between serotypes. These include looking for the presence or absence of antigens, phage typing, molecul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are part of the normal microbiota of the gut, where they constitute about 0.1%, along with other facultative anaerobes. These bacteria are mostly harmless or even beneficial to humans. For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by harmful pathogenic bacteria. These mutually beneficial relationships between ''E. coli'' and humans are a type of mutualistic biological relationship—where both the humans and the ''E. coli'' are benefitting each other. ''E. coli'' is expelled into the environment within fecal matter. The bacterium grows massi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus Anthracis
''Bacillus anthracis'' is a gram-positive and rod-shaped bacterium that causes anthrax, a deadly disease to livestock and, occasionally, to humans. It is the only permanent (obligate) pathogen within the genus ''Bacillus''. Its infection is a type of zoonosis, as it is transmitted from animals to humans. It was discovered by a German physician Robert Koch in 1876, and became the first bacterium to be experimentally shown as a pathogen. The discovery was also the first scientific evidence for the germ theory of diseases. ''B. anthracis'' measures about 3 to 5 μm long and 1 to 1.2 μm wide. The reference genome consists of a 5,227,419 bp circular chromosome and two extrachromosomal DNA plasmids, pXO1 and pXO2, of 181,677 and 94,830 bp respectively, which are responsible for the pathogenicity. It forms a protective layer called endospore by which it can remain inactive for many years and suddenly becomes infective under suitable environmental conditions. Because of the resilience ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Typhlosole
In Morphology (biology), biological morphology, a typhlosole is an internal fold of the Gut (anatomy), intestine or intestine inner wall. Typhlosoles occur in bivalve mollusks, lampreys and some annelids and echinoderms. In earthworms, it is a dorsal flap of the intestine that runs along most of its length, effectively forming a tube within a tube, and increasing the absorption area by that of its inner surface. Its function is to increase intestine surface area for more efficient absorption of digested nutrients. In different earthworm families, the typhlosole appears to have multiple origins. The Lumbricidae, for example, have a typhlosole which is an infolding of all layers of the intestine wall, whereas in some other families (e.g. Megascolecidae), it is an infolding of only the inner layer, and in many earthworms it is absent. In shipworms, the typhlosole is the organ where the lignin in wood are digested by Symbiosis, symbiont bacteria of the "Alteromonas, Alteromonas-like ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
APOBEC
image:Apobec.J.Steinfeld.D.png, 300px, upExample of a member of the APOBEC family, APOBEC-2. A cytidine deaminase from ''Homo sapiens''.; ; rendered usinPyMOL APOBEC ("apolipoprotein B mRNA editing enzyme, catalytic polypeptide") is a family of evolutionarily conserved cytidine deaminases. Function A mechanism of generating protein diversity is mRNA editing. The APOBEC family of proteins perform mRNA modifications by deaminating cytidine bases to uracil. The N-terminal domain of APOBEC-like proteins is the catalytic domain, while the C-terminal domain is a pseudocatalytic domain. More specifically, the catalytic domain is a zinc dependent cytidine deaminase domain and is essential for cytidine deamination. The positively charged zinc ion in the catalytic domain attracts to the partial-negative charge of RNA. In the case of APOBEC-1, the mRNA transcript of intestinal apolipoprotein B is altered. RNA editing by APOBEC-1 requires homodimerization and this complex interacts wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytidine Deaminase
Cytidine deaminase is an enzyme that in humans is encoded by the ''CDA'' gene. This gene encodes an enzyme involved in pyrimidine salvaging. The encoded protein forms a homotetramer that catalyzes the irreversible hydrolytic deamination of cytidine and deoxycytidine to uridine and deoxyuridine, respectively. It is one of several deaminases responsible for maintaining the cellular pyrimidine pool. Mutations in this gene are associated with decreased sensitivity to the cytosine nucleoside analogue cytosine arabinoside used in the treatment of certain childhood leukemias. Most cytidine deaminases act on RNA, and the few that act on DNA require ssDNA. A related activation-induced (cytidine) deaminase (AID) regulates antibody diversification, especially the process of somatic hypermutation Somatic hypermutation (or SHM) is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it (e.g. microbes). A major component of the process of affinity ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |