Protein targeting on:
[Wikipedia]
[Google]
[Amazon]
:''This article deals with protein targeting in
 In 1970,
In 1970,
 Most secretory and membrane-bound proteins are co-translationally translocated. Proteins that reside in the
Most secretory and membrane-bound proteins are co-translationally translocated. Proteins that reside in the
 While some proteins in the mitochondria originate from
While some proteins in the mitochondria originate from
 Proteins targeted to the mitochondrial matrix first involves interactions between the matrix targeting sequence located at the N-terminus and the outer membrane import receptor complex TOM20/22. In addition to the docking of internal sequences and
Proteins targeted to the mitochondrial matrix first involves interactions between the matrix targeting sequence located at the N-terminus and the outer membrane import receptor complex TOM20/22. In addition to the docking of internal sequences and

 Peroxisomes contain a single phospholipid bilayer that surrounds the peroxisomal matrix containing a wide variety of proteins and enzymes that participate in anabolism and catabolism. Since it contains no internal DNA like that of the mitochondria or chloroplast all peroxisomal proteins are encoded by nuclear genes. To date there are two types of known Peroxisome Targeting Signals (PTS):
# Peroxisome targeting signal 1 (PTS1): a C-terminal tripeptide with a consensus sequence (S/A/C)-(K/R/H)-(L/A). The most common PTS1 is serine- lysine- leucine (SKL). The initial research that led to the discovery of this consensus observed that when firefly luciferase was expressed in cultured insect cells it was targeted to the peroxisome. By testing a variety of mutations in the gene encoding the expressed luciferase, the consensus sequence was then determined. It has also been found that by adding this C-terminal sequence of SKL to a cytosolic protein that it becomes targeted for transport to the peroxisome. The majority of peroxisomal matrix proteins possess this PTS1 type signal.
# Peroxisome targeting signal 2 (PTS2): a nonapeptide located near the N-terminus with a consensus sequence (R/K)-(L/V/I)-XXXXX-(H/Q)-(L/A/F) (where X can be any amino acid).
There are also proteins that possess neither of these signals. Their transport may be based on a so-called "piggy-back" mechanism: such proteins associate with PTS1-possessing matrix proteins and are translocated into the peroxisomal matrix together with them.
In the case of cytosolic proteins that are produced with the PTS1 C-terminal sequence, its path to the peroxisomal matrix is dependent upon binding to another cytosolic protein called pex5 (peroxin 5). Once bound, pex5 interacts with a peroxisomal membrane protein
Peroxisomes contain a single phospholipid bilayer that surrounds the peroxisomal matrix containing a wide variety of proteins and enzymes that participate in anabolism and catabolism. Since it contains no internal DNA like that of the mitochondria or chloroplast all peroxisomal proteins are encoded by nuclear genes. To date there are two types of known Peroxisome Targeting Signals (PTS):
# Peroxisome targeting signal 1 (PTS1): a C-terminal tripeptide with a consensus sequence (S/A/C)-(K/R/H)-(L/A). The most common PTS1 is serine- lysine- leucine (SKL). The initial research that led to the discovery of this consensus observed that when firefly luciferase was expressed in cultured insect cells it was targeted to the peroxisome. By testing a variety of mutations in the gene encoding the expressed luciferase, the consensus sequence was then determined. It has also been found that by adding this C-terminal sequence of SKL to a cytosolic protein that it becomes targeted for transport to the peroxisome. The majority of peroxisomal matrix proteins possess this PTS1 type signal.
# Peroxisome targeting signal 2 (PTS2): a nonapeptide located near the N-terminus with a consensus sequence (R/K)-(L/V/I)-XXXXX-(H/Q)-(L/A/F) (where X can be any amino acid).
There are also proteins that possess neither of these signals. Their transport may be based on a so-called "piggy-back" mechanism: such proteins associate with PTS1-possessing matrix proteins and are translocated into the peroxisomal matrix together with them.
In the case of cytosolic proteins that are produced with the PTS1 C-terminal sequence, its path to the peroxisomal matrix is dependent upon binding to another cytosolic protein called pex5 (peroxin 5). Once bound, pex5 interacts with a peroxisomal membrane protein
Phobius
predicts signal peptides based on a supplied primary sequence.
SignalP
predicts signal peptide cleavage sites.
LOCtree
predicts the subcellular localization of proteins.
eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
s unless specified otherwise.''
Protein targeting or protein sorting is the biological mechanism by which protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion 440px
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classic ...
. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
History
Günter Blobel
Günter Blobel (; May 21, 1936 – February 18, 2018) was a Silesian German and American biologist and 1999 Nobel Prize laureate in Physiology for the discovery that proteins have intrinsic signals that govern their transport and localization in ...
conducted experiments on protein translocation across membranes
A membrane is a selective barrier; it allows some things to pass through but stops others. Such things may be molecules, ions, or other small particles. Membranes can be generally classified into synthetic membranes and biological membranes. Bi ...
. Blobel, then an assistant professor at Rockefeller University, built upon the work of his colleague George Palade
George Emil Palade (; November 19, 1912 – October 7, 2008) was a Romanian cell biologist. Described as "the most influential cell biologist ever",
. Palade had previously demonstrated that non-secreted proteins were translated by free ribosomes in the cytosol, while secreted proteins (and target proteins, in general) were translated by ribosomes bound to the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ...
. Candidate explanations at the time postulated a processing difference between free and ER-bound ribosomes, but Blobel hypothesized that protein targeting relied on characteristics inherent to the proteins, rather than a difference in ribosomes. Supporting his hypothesis, Blobel discovered that many proteins have a short amino acid sequence at one end that functions like a postal code specifying an intracellular or extracellular destination. He described these short sequences (generally 13 to 36 amino acids residues) as signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-te ...
s or signal sequences and was awarded the 1999 Nobel prize in Physiology for the same.
Signal peptides
Signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-te ...
s serve as targeting signals, enabling cellular transport machinery to direct proteins to specific intracellular or extracellular locations. While no consensus sequence has been identified for signal peptides, many nonetheless possess a characteristic tripartite structure:
# A positively charged, hydrophilic region near the N-terminal.
# A span of 10 to 15 hydrophobic amino acids near the middle of the signal peptide.
# A slightly polar region near the C-terminal, typically favoring amino acids with smaller side chains at positions approaching the cleavage site.
After a protein has reached its destination, the signal peptide is generally cleaved by a signal peptidase. Consequently, most mature proteins do not contain signal peptides. While most signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-te ...
s are found at the N-terminal, in peroxisomes the targeting sequence is located on the C-terminal extension. Unlike signal peptides, signal patches are composed by amino acid residues that are discontinuous in the primary sequence
Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length ...
but become functional when folding brings them together on the protein surface. Unlike most signal sequences, signal patches are not cleaved after sorting is complete. In addition to intrinsic signaling sequences, protein modifications like glycosylation can also induce targeting to specific intracellular or extra cellular regions.
Protein translocation
Since thetranslation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
of mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
into protein by a ribosome takes place within the cytosol, proteins destined for secretion or a specific organelle must be translocated. This process can occur during translation, known as co-translational translocation, or after translation is complete, known as post-translational translocation.
Co-translational translocation
 Most secretory and membrane-bound proteins are co-translationally translocated. Proteins that reside in the
Most secretory and membrane-bound proteins are co-translationally translocated. Proteins that reside in the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ...
(ER), golgi or endosomes also use the co-translational translocation pathway. This process begins while the protein is being synthesized on the ribosome, when a signal recognition particle
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein ( protein- RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma memb ...
(SRP) recognizes an N-terminal signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-te ...
of the nascent protein. Binding of the SRP temporarily pauses synthesis while the ribosome-protein complex is transferred to an SRP receptor on the ER in eukaryotes, and the plasma membrane in prokaryotes. There, the nascent protein is inserted into the translocon, a membrane-bound protein conducting channel composed of the Sec61 translocation complex in eukaryotes, and the homologous SecYEG complex in prokaryotes. In secretory proteins and type I transmembrane proteins, the signal sequence is immediately cleaved from the nascent polypeptide once it has been translocated into the membrane of the ER (eukaryotes) or plasma membrane (prokaryotes) by signal peptidase. The signal sequence of type II membrane proteins and some polytopic membrane proteins are not cleaved off and therefore are referred to as signal anchor sequences. Within the ER, the protein is first covered by a chaperone protein
In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to as ...
to protect it from the high concentration of other proteins in the ER, giving it time to fold correctly. Once folded, the protein is modified as needed (for example, by glycosylation), then transported to the Golgi for further processing and goes to its target organelles or is retained in the ER by various ER retention mechanisms.
The amino acid chain of transmembrane proteins, which often are transmembrane receptors
Cell surface receptors (membrane receptors, transmembrane receptors) are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving (binding to) extracellular molecules. They are specialized integral m ...
, passes through a membrane one or several times. These proteins are inserted into the membrane by translocation, until the process is interrupted by a stop-transfer sequence, also called a membrane anchor or signal-anchor sequence. These complex membrane proteins are currently characterized using the same model of targeting that has been developed for secretory proteins. However, many complex multi-transmembrane proteins contain structural aspects that do not fit this model. Seven transmembrane G-protein coupled receptors (which represent about 5% of the genes in humans) mostly do not have an amino-terminal signal sequence. In contrast to secretory proteins, the first transmembrane domain acts as the first signal sequence, which targets them to the ER membrane. This also results in the translocation of the amino terminus of the protein into the ER membrane lumen. This translocation, which has been demonstrated with opsin
Animal opsins are G-protein-coupled receptors and a group of proteins made light-sensitive via a chromophore, typically retinal. When bound to retinal, opsins become Retinylidene proteins, but are usually still called opsins regardless. Most ...
with in vitro experiments, breaks the usual pattern of "co-translational" translocation which has always held for mammalian proteins targeted to the ER. A great deal of the mechanics of transmembrane topology and folding remains to be elucidated.
Post-translational translocation
Even though most secretory proteins are co-translationally translocated, some are translated in the cytosol and later transported to the ER/plasma membrane by a post-translational system. In prokaryotes this process requires certain cofactors such as SecA and SecB and is facilitated by Sec62 and Sec63, two membrane-bound proteins. The Sec63 complex, which is embedded in the ER membrane, causes hydrolysis of ATP, allowing chaperone proteins to bind to an exposed peptide chain and slide the polypeptide into the ER lumen. Once in the lumen the polypeptide chain can be folded properly. This process only occurs in unfolded proteins located in the cytosol. In addition, proteins targeted to other cellular destinations, such asmitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
, chloroplasts, or peroxisomes, use specialized post-translational pathways. Proteins targeted for the nucleus are also translocated post-translationally through the addition of a nuclear localization signal (NLS) that promotes passage through the nuclear envelope via nuclear pore
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore comple ...
s.
Sorting of proteins
Mitochondria

 While some proteins in the mitochondria originate from
While some proteins in the mitochondria originate from mitochondrial DNA
Mitochondrial DNA (mtDNA or mDNA) is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, such as adenosine triphosphate (ATP). Mitochondrial D ...
within the organelle, most mitochondrial proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
are synthesized as cytosolic
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrio ...
precursors containing uptake peptide signals. Unfolded proteins bound by cytosolic
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrio ...
chaperone hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an import ...
that are targeted to the mitochondria may be localized to four different areas depending on their sequences. They may be targeted to the mitochondrial matrix, the outer membrane, the intermembrane space, or the inner membrane. Defects in any one or more of these processes has been linked to health and disease.
Mitochondrial Matrix
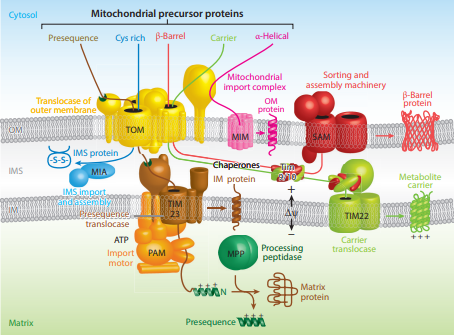
 Proteins targeted to the mitochondrial matrix first involves interactions between the matrix targeting sequence located at the N-terminus and the outer membrane import receptor complex TOM20/22. In addition to the docking of internal sequences and
Proteins targeted to the mitochondrial matrix first involves interactions between the matrix targeting sequence located at the N-terminus and the outer membrane import receptor complex TOM20/22. In addition to the docking of internal sequences and cytosolic
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrio ...
chaperones to TOM70. Where TOM is an abbreviation for translocase of the outer membrane. Binding of the matrix targeting sequence to the import receptor triggers a handoff of the polypeptide to the general import core (GIP) known as TOM40. The general import core (TOM40) then feeds the polypeptide chain through the intermembrane space and into another translocase complex TIM17/23/44 located on the inner mitochondrial membrane. This is accompanied by the necessary release of the cytosolic
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrio ...
chaperones that maintain an unfolded state prior to entering the mitochondria. As the polypeptide enters the matrix, the signal sequence is cleaved by a processing peptidase and the remaining sequences are bound by mitochondrial chaperones to await proper folding and activity. The push and pull of the polypeptide from the cytosol to the intermembrane space and then the matrix is achieved by an electrochemical gradient that is established by the mitochondrion during oxidative phosphorylation
Oxidative phosphorylation (UK , US ) or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine t ...
. In which a mitochondrion active in metabolism
Metabolism (, from el, μεταβολή ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run ...
has generated a negative potential inside the matrix and a positive potential in the intermembrane space. It is this negative potential inside the matrix that directs the positively charged regions of the targeting sequence into its desired location.
Mitochondrial Inner Membrane
Targeting of mitochondrial proteins to the inner membrane may follow 3 different pathways depending upon their overall sequences, however, entry from the outer membrane remains the same using the import receptor complex TOM20/22 and TOM40 general import core. The first pathway for proteins targeted to the inner membrane follows the same steps as those designated to the matrix where it contains a matrix targeting sequence that channels the polypeptide to the inner membrane complex containing the previously mentioned translocase complex TIM17/23/44. However, the difference is that the peptides that are designated to the inner membrane and not the matrix contain an upstream sequence called the stop-transfer-anchor sequence. This stop-transfer-anchor sequence is a hydrophobic region that embeds itself into the phospholipid bilayer of the inner membrane and prevents translocation further into the mitochondrion. The second pathway for proteins targeted to the inner membrane follows the matrix localization pathway in its entirety. However, instead of a stop-transfer-anchor sequence, it contains another sequence that interacts with an inner membrane protein called Oxa-1 once inside the matrix that will embed it into the inner membrane. The third pathway for mitochondrial proteins targeted to the inner membrane follow the same entry as the others into the outer membrane, however, this pathway utilizes the translocase complex TIM22/54 assisted by complex TIM9/10 in the intermembrane space to anchor the incoming peptide into the membrane. The peptides for this last pathway do not contain a matrix targeting sequence, but instead contain several internal targeting sequences.Mitochondrial Intermembrane Space
If instead the precursor protein is designated to the intermembrane space of the mitochondrion, there are two pathways this may occur depending on the sequences being recognized. The first pathway to the intermembrane space follows the same steps for an inner membrane targeted protein. However, once bound to the inner membrane the C-terminus of the anchored protein is cleaved via a peptidase that liberates the preprotein into the intermembrane space so it can fold into its active state. One of the greatest examples for a protein that follows this pathway is cytochrome b2, that upon being cleaved will interact with a heme cofactor and become active. The second intermembrane space pathway does not utilize any inner membrane complexes and therefor does not contain a matrix targeting signal. Instead, it enters through the general import core TOM40 and is further modified in the intermembrane space to achieve its active conformation. TIM9/10 is an example of a protein that follows this pathway in order to be in the location it needs to be to assist in inner membrane targeting.Mitochondrial Outer Membrane
Outer membrane targeting simply involves the interaction of precursor proteins with the outer membrane translocase complexes that embeds it into the membrane via internal-targeting sequences that are to form hydrophobic alpha helices or beta barrels that span the phospholipid bilayer. This may occur by two different routes depending on the preprotein internal sequences. If the preprotein contains internal hydrophobic regions capable of forming alpha helices, then the preprotein will utilize the mitochondrial import complex (MIM) and be transferred laterally to the membrane. For preproteins containing hydrophobic internal sequences that correlate to beta-barrel forming proteins, they will be imported from the aforementioned outer membrane complex TOM20/22 to the intermembrane space. In which they will interact with TIM9/10 intermembrane-space protein complex that transfers them tosorting and assembly machinery
The outer mitochondrial membrane is made up of two essential proteins, Tom40 and Sam50.
Tom40
Tom40 is a protein import pore required for the import of precursor proteins across the outer mitochondrial membrane, and it makes up part of the tran ...
(SAM) that is present in the outer membrane that laterally displaces the targeted protein as a beta-barrel.
Chloroplasts
Chloroplasts are similar to mitochondria in that they contain their own DNA for production of some of their components. However, the majority of their proteins are obtained via post-translational translocation and arise from nuclear genes. Proteins may be targeted to several sites of the chloroplast depending on their sequences such as the outer envelope, inner envelope, stroma, thylakoid lumen, or the thylakoid membrane. Proteins targeted to the envelope of chloroplasts usually lack cleavable sorting sequence and are laterally displaced via membrane sorting complexes. General import for the majority of preproteins requires translocation from the cytosol through the Toc and Tic complexes located within the chloroplast envelope. Where Toc is an abbreviation for the translocase of the outer chloroplast envelope and Tic is the translocase of the inner chloroplast envelope. There is a minimum of three proteins that make up the function of the Toc complex. Two of which, referred to as Toc159 and Toc34, are responsible for the docking of stromal import sequences and both contain GTPase activity. The third known as Toc 75, is the actual translocation channel that feeds the recognized preprotein by Toc159/34 into the chloroplast.Stroma
Targeting to the stroma requires the preprotein to have a stromal import sequence that is recognized by the Tic complex of the inner envelope upon being translocated from the outer envelope by the Toc complex. The Tic complex is composed of at least five different Tic proteins that are required to form the translocation channel across the inner envelope. Upon being delivered to the stroma, the stromal import sequence is cleaved off via a signal peptidase. This delivery process to the stroma is currently known to be driven by ATP hydrolysis via stromal HSP chaperones, instead of the transmembrane electrochemical gradient that is established in mitochondria to drive protein import. Further intra-chloroplast sorting depends on additional target sequences such as those designated to the thylakoid membrane or the thylakoid lumen.Thylakoid Lumen
If a protein is to be targeted to the thylakoid lumen, this may occur via four differently known routes that closely resemble bacterial protein transport mechanisms. The route that is taken depends upon the protein delivered to the stroma being in either an unfolded or metal-bound folded state. Both of which will still contain a thylakoid targeting sequence that is also cleaved upon entry to the lumen. While protein import into the stroma is ATP-driven, the pathway for metal-bound proteins in a folded state to the thylakoid lumen has been shown to be driven by a pH gradient.
Thylakoid Membrane
Proteins bound for the membrane of the thylakoid will follow up to four known routes that are illustrated in the corresponding figure shown. They may follow a co-translational insertion route that utilizes stromal ribosomes and the SecY/E transmembrane complex, the SRP-dependent pathway, the spontaneous insertion pathway, or the GET pathway. The last of the three are post-translational pathways originating from nuclear genes and therefor constitute the majority of proteins targeted to the thylakoid membrane. According to recent review articles in the journal of biochemistry and molecular biology, the exact mechanisms are not yet fully understood.Both chloroplasts and mitochondria
Many proteins are needed in bothmitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
and chloroplasts. In general the dual-targeting peptide is of intermediate character to the two specific ones. The targeting peptides of these protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s have a high content of basic and hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water.
Hydrophobic molecules tend to be nonpolar and, ...
amino acids, a low content of negatively charged amino acids. They have a lower content of alanine and a higher content of leucine and phenylalanine. The dual targeted proteins have a more hydrophobic targeting peptide than both mitochondrial and chloroplastic ones. However, it is tedious to predict if a peptide is dual-targeted or not based on its physio-chemical characteristics.
Peroxisomes
 Peroxisomes contain a single phospholipid bilayer that surrounds the peroxisomal matrix containing a wide variety of proteins and enzymes that participate in anabolism and catabolism. Since it contains no internal DNA like that of the mitochondria or chloroplast all peroxisomal proteins are encoded by nuclear genes. To date there are two types of known Peroxisome Targeting Signals (PTS):
# Peroxisome targeting signal 1 (PTS1): a C-terminal tripeptide with a consensus sequence (S/A/C)-(K/R/H)-(L/A). The most common PTS1 is serine- lysine- leucine (SKL). The initial research that led to the discovery of this consensus observed that when firefly luciferase was expressed in cultured insect cells it was targeted to the peroxisome. By testing a variety of mutations in the gene encoding the expressed luciferase, the consensus sequence was then determined. It has also been found that by adding this C-terminal sequence of SKL to a cytosolic protein that it becomes targeted for transport to the peroxisome. The majority of peroxisomal matrix proteins possess this PTS1 type signal.
# Peroxisome targeting signal 2 (PTS2): a nonapeptide located near the N-terminus with a consensus sequence (R/K)-(L/V/I)-XXXXX-(H/Q)-(L/A/F) (where X can be any amino acid).
There are also proteins that possess neither of these signals. Their transport may be based on a so-called "piggy-back" mechanism: such proteins associate with PTS1-possessing matrix proteins and are translocated into the peroxisomal matrix together with them.
In the case of cytosolic proteins that are produced with the PTS1 C-terminal sequence, its path to the peroxisomal matrix is dependent upon binding to another cytosolic protein called pex5 (peroxin 5). Once bound, pex5 interacts with a peroxisomal membrane protein
Peroxisomes contain a single phospholipid bilayer that surrounds the peroxisomal matrix containing a wide variety of proteins and enzymes that participate in anabolism and catabolism. Since it contains no internal DNA like that of the mitochondria or chloroplast all peroxisomal proteins are encoded by nuclear genes. To date there are two types of known Peroxisome Targeting Signals (PTS):
# Peroxisome targeting signal 1 (PTS1): a C-terminal tripeptide with a consensus sequence (S/A/C)-(K/R/H)-(L/A). The most common PTS1 is serine- lysine- leucine (SKL). The initial research that led to the discovery of this consensus observed that when firefly luciferase was expressed in cultured insect cells it was targeted to the peroxisome. By testing a variety of mutations in the gene encoding the expressed luciferase, the consensus sequence was then determined. It has also been found that by adding this C-terminal sequence of SKL to a cytosolic protein that it becomes targeted for transport to the peroxisome. The majority of peroxisomal matrix proteins possess this PTS1 type signal.
# Peroxisome targeting signal 2 (PTS2): a nonapeptide located near the N-terminus with a consensus sequence (R/K)-(L/V/I)-XXXXX-(H/Q)-(L/A/F) (where X can be any amino acid).
There are also proteins that possess neither of these signals. Their transport may be based on a so-called "piggy-back" mechanism: such proteins associate with PTS1-possessing matrix proteins and are translocated into the peroxisomal matrix together with them.
In the case of cytosolic proteins that are produced with the PTS1 C-terminal sequence, its path to the peroxisomal matrix is dependent upon binding to another cytosolic protein called pex5 (peroxin 5). Once bound, pex5 interacts with a peroxisomal membrane protein pex14
Peroxisomal membrane protein PEX14 is a protein that in humans is encoded by the ''PEX14'' gene.
Function
This gene encodes an essential component of the peroxisomal import machinery. The protein is integrated into peroxisome membranes with i ...
to form a complex. When the pex5 protein with bound cargo interacts with the pex14 membrane protein, the complex induces the release of the targeted protein into the matrix. Upon releasing the cargo protein into the matrix, pex5 dissociation from pex14 occurs via ubiquitinylation by a membrane complex comprising pex2, pex12, and pex10 followed by an ATP dependent removal involving the cytosolic protein complex pex1 and pex6. The cycle for pex5 mediated import into the peroxisomal matrix is restored after the ATP dependent removal of ubiquitin and is free to bind with another protein containing a PTS1 sequence. Proteins containing a PTS2 targeting sequence are mediated by a different cytosolic protein but are believed to follow a similar mechanism to that of those containing the PTS1 sequence.
Diseases
Protein transport is defective in the following genetic diseases: * Mohr–Tranebjaerg syndrome * Zellweger syndrome *Adrenoleukodystrophy
Adrenoleukodystrophy (ALD) is a disease linked to the X chromosome. It is a result of fatty acid buildup caused by peroxisomal fatty acid beta oxidation which results in the accumulation of very long chain fatty acids in tissues throughout the ...
(ALD).
*Refsum disease
Refsum disease is an autosomal recessive neurological disease that results in the over-accumulation of phytanic acid in cells and tissues. It is one of several disorders named after Norwegian neurologist Sigvald Bernhard Refsum (1907–1991). Ref ...
*Parkinson's disease
Parkinson's disease (PD), or simply Parkinson's, is a long-term degenerative disorder of the central nervous system that mainly affects the motor system. The symptoms usually emerge slowly, and as the disease worsens, non-motor symptoms beco ...
* Hypercholesterolemia, atherosclerosis, obesity
Obesity is a medical condition, sometimes considered a disease, in which excess body fat has accumulated to such an extent that it may negatively affect health. People are classified as obese when their body mass index (BMI)—a person's ...
, and diabetes
Diabetes, also known as diabetes mellitus, is a group of metabolic disorders characterized by a high blood sugar level ( hyperglycemia) over a prolonged period of time. Symptoms often include frequent urination, increased thirst and increased ...
In bacteria and archaea
As discussed above (see protein translocation), most prokaryotic membrane-bound and secretory proteins are targeted to the plasma membrane by either a co-translation pathway that uses bacterial SRP or a post-translation pathway that requires SecA and SecB. At the plasma membrane, these two pathways deliver proteins to the SecYEG translocon for translocation. Bacteria may have a single plasma membrane ( Gram-positive bacteria), or an inner membrane plus an outer membrane separated by the periplasm ( Gram-negative bacteria). Besides the plasma membrane the majority of prokaryotes lack membrane-bound organelles as found in eukaryotes, but they may assemble proteins onto various types of inclusions such as gas vesicles and storage granules.Gram-negative bacteria
In gram-negative bacteria proteins may be incorporated into the plasma membrane, the outer membrane, the periplasm or secreted into the environment. Systems for secreting proteins across the bacterial outer membrane may be quite complex and play key roles in pathogenesis. These systems may be described as type I secretion, type II secretion, etc.Gram-positive bacteria
In most gram-positive bacteria, certain proteins are targeted for export across the plasma membrane and subsequent covalent attachment to the bacterial cell wall. A specialized enzyme, sortase, cleaves the target protein at a characteristic recognition site near the protein C-terminus, such as an LPXTG motif (where X can be any amino acid), then transfers the protein onto the cell wall. Several analogous systems are found that likewise feature a signature motif on the extra-cytoplasmic face, a C-terminal transmembrane domain, and cluster of basic residues on the cytosolic face at the protein's extreme C-terminus. The PEP-CTERM/ exosortase system, found in many Gram-negative bacteria, seems to be related toextracellular polymeric substance
Extracellular polymeric substances (EPSs) are natural polymers of high molecular weight secreted by microorganisms into their environment. EPSs establish the functional and structural integrity of biofilms, and are considered the fundamental comp ...
production. The PGF-CTERM/archaeosortase A system in archaea is related to S-layer production. The GlyGly-CTERM/rhombosortase system, found in the Shewanella, Vibrio, and a few other genera, seems involved in the release of proteases, nucleases, and other enzymes.
Bioinformatic tools
*Minimotif Miner Minimotif Miner is a program and database designed to identify minimotifs in any protein. Minimotifs are short contiguous peptide sequences that are known to have a function in at least one protein. Minimotifs are also called sequence motifs or sho ...
is a bioinformatics tool that searches protein sequence queries for a known protein targeting sequence motifs.
Phobius
predicts signal peptides based on a supplied primary sequence.
SignalP
predicts signal peptide cleavage sites.
LOCtree
predicts the subcellular localization of proteins.
See also
*Bulk flow
Mass flow, also known as mass transfer and bulk flow, is the movement of fluids down a pressure or temperature gradient,Moyes & Schulte (2008). Principles of Animal Physiology. Pearson Benjamin Cummings. San Francisco, California particularly in ...
* COPI
* COPII
The Coat Protein Complex II, or COPII, is a group of proteins that facilitate the formation of vesicles to transport proteins from the endoplasmic reticulum to the Golgi apparatus or endoplasmic-reticulum–Golgi intermediate compartment. This ...
* Clathrin
* LocDB
LocDB is an expert-curated database that collects experimental annotations for the subcellular localization of proteins in Homo sapiens (human) and Arabidopsis thaliana (Weed). The database also contains predictions of subcellular localization from ...
* PSORTdb
* Signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-te ...
References
External links
* {{DEFAULTSORT:Protein Targeting Post-translational modification Membrane proteins