LSm on:
[Wikipedia]
[Google]
[Amazon]
 In
In
 Uridine phosphate binds in archaeal Sm1 between the β2b/β3a loop and β4b/β5 loop. The
Uridine phosphate binds in archaeal Sm1 between the β2b/β3a loop and β4b/β5 loop. The
Pfam entry LSM. Pfam is the Sanger Institute database, which is a collection of protein families and domains.
{{DEFAULTSORT:Lsm Protein families Spliceosome
molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and phys ...
, LSm proteins are a family of RNA-binding protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s found in virtually every cellular organism
In biology, an organism () is any living system that functions as an individual entity. All organisms are composed of cells ( cell theory). Organisms are classified by taxonomy into groups such as multicellular animals, plants, and fu ...
. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be ...
were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecule
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and b ...
s, and play a large number of various roles in mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
processing and regulation.
The Sm proteins were first discovered as antigens
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. ...
targeted by so-called anti-Sm antibodies in a patient with a form of systemic lupus erythematosus
Lupus, technically known as systemic lupus erythematosus (SLE), is an autoimmune disease in which the body's immune system mistakenly attacks healthy tissue in many parts of the body. Symptoms vary among people and may be mild to severe. Commo ...
(SLE), a debilitating autoimmune disease
An autoimmune disease is a condition arising from an abnormal immune response to a functioning body part. At least 80 types of autoimmune diseases have been identified, with some evidence suggesting that there may be more than 100 types. Nearly a ...
. They were named Sm proteins in honor of Stephanie Smith, a patient who suffered from SLE. Other proteins with very similar structures were subsequently discovered and named LSm proteins. New members of the LSm protein family continue to be identified and reported.
Proteins with similar structures are grouped into a hierarchy of protein families, superfamilies, and folds. The LSm protein structure is an example of a small beta sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a ge ...
folded into a short barrel. Individual LSm proteins assemble into a six or seven member doughnut ring (more properly termed a torus
In geometry, a torus (plural tori, colloquially donut or doughnut) is a surface of revolution generated by revolving a circle in three-dimensional space about an axis that is coplanar with the circle.
If the axis of revolution does n ...
), which usually binds to a small RNA
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding. RNA silencing is often a function of these molecules, with the most common and well-studied example being RNA interference ( ...
molecule to form a ribonucleoprotein complex. The LSm torus assists the RNA molecule to assume and maintain its proper three-dimensional structure. Depending on which LSm proteins and RNA molecule are involved, this ribonucleoprotein complex facilitates a wide variety of RNA processing including degradation, editing, splicing, and regulation.
Alternate terms for LSm family are LSm fold and Sm-like fold, and alternate capitalization styles such as lsm, LSM, and Lsm are common and equally acceptable.
History
Discovery of the Smith antigen
The story of the discovery of the first LSmprotein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s begins with a young woman, Stephanie Smith, who was diagnosed in 1959 with systemic lupus erythematosus (SLE), eventually succumbing to complications of the disease in 1969 at the age of 22. During this period, she was treated at New York's Rockefeller University
The Rockefeller University is a private biomedical research and graduate-only university in New York City, New York. It focuses primarily on the biological and medical sciences and provides doctoral and postdoctoral education. It is classif ...
Hospital, under the care of Dr. Henry Kunkel and Dr. Eng Tan. As those with an autoimmune disease
An autoimmune disease is a condition arising from an abnormal immune response to a functioning body part. At least 80 types of autoimmune diseases have been identified, with some evidence suggesting that there may be more than 100 types. Nearly a ...
, SLE patients produce antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of ...
to antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune respon ...
s in their cells' nuclei, most frequently to their own DNA. However, Dr. Kunkel and Dr. Tan found in 1966 that Ms. Smith produced antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of ...
to a set of nuclear proteins, which they named the 'smith antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune respon ...
' (Sm Ag). About 30% of SLE patients produce antibodies to these proteins, as opposed to double stranded DNA. This discovery improved diagnostic testing for SLE, but the nature and function of this antigen was unknown.
Sm proteins, snRNPs, the spliceosome and messenger RNA splicing
Research continued during the 1970s and early 1980s. The smith antigen was found to be a complex of ribonucleic acid ( RNA) molecules and multiple proteins. A set of uridine-richsmall nuclear RNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribe ...
(snRNA) molecules was part of this complex, and given the names U1, U2, U4, U5 and U6. Four of these snRNAs (U1, U2, U4 and U5) were found to be tightly bound to several small proteins, which were named SmB, SmD, SmE, SmF, and SmG in decreasing order of size. SmB has an alternatively spliced variant, SmB', and a very similar protein, SmN, replaces SmB'/B in certain (mostly neural) tissues. SmD was later discovered to be a mixture of three proteins, which were named SmD1, SmD2 and SmD3. These nine proteins (SmB, SmB', SmN, SmD1, SmD2, SmD3, SmE, SmF and SmG) became known as the Sm core proteins, or simply Sm proteins. The snRNAs are complexed with the Sm core proteins and with other proteins to form particles in the cell's nucleus called small nuclear ribonucleoproteins, or snRNPs. By the mid 1980s, it became clear that these snRNPs help form a large (4.8 MD molecular weight
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and bio ...
) complex, called the spliceosome, around pre-mRNA, excising portions of the pre-mRNA called intron
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of ...
s and splicing the coding portions (exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequen ...
s) together. After a few more modifications, the spliced pre-mRNA becomes messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) which is then exported from the nucleus and translated
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
into a protein by ribosomes.
Discovery of proteins similar to the Sm proteins
The snRNA U6 (unlike U1, U2, U4 and U5) does not associate with the Sm proteins, even though the U6 snRNP is a central component in the spliceosome. In 1999 a protein heteromer was found that binds specifically to U6, and consisted of seven proteins clearly homologous to the Sm proteins. These proteins were denoted LSm (like Sm) proteins (LSm1, LSm2, LSm3, LSm4, LSm5, LSm6 and LSm7), with the similar LSm8 protein identified later. In the bacterium ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'', the Sm-like protein HF-I encoded by the gene ''hfq
The Hfq protein (also known as HF-I protein) encoded by the ''hfq'' gene was discovered in 1968 as an ''Escherichia coli'' host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RN ...
'' was described in 1968 as an essential host factor for RNA bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bac ...
Qβ replication. The genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
of ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have b ...
'' (Baker's Yeast) was sequenced in the mid-1990s, providing a rich resource for identifying homologs of these human proteins. Subsequently, as more eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
s genomes were sequenced, it became clear that eukaryotes, in general, share homologs to the same set of seven Sm and eight LSm proteins. Soon after, proteins homologous to these eukaryote LSm proteins were found in Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaeba ...
(Sm1 and Sm2) and Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
(Hfq and YlxS homologs). The archaeal LSm proteins are more similar to the eukaryote LSm proteins than either are to bacterial LSm proteins. The LSm proteins described thus far were rather small proteins, varying from 76 amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
s (8.7 kD molecular weight
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and bio ...
) for human SmG to 231 amino acids (29 kD molecular weight) for human SmB. But recently, larger proteins have been discovered that include a LSm structural domain in addition to other protein structural domains (such as LSm10, LSm11, LSm12, LSm13, LSm14, LSm15, LSm16, ataxin-2, as well as archaeal Sm3).
Discovery of the LSm fold
Around 1995, comparisons between the various LSm homologs identified twosequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ' ...
s, 32 nucleic acids long (14 amino acids), that were very similar in each LSm homolog, and were separated by a non-conserved region of variable length. This indicated the importance of these two sequence motifs (named Sm1 and Sm2), and suggested that all LSm protein genes evolved from a single ancestral gene. In 1999, crystals of recombinant Sm proteins were prepared, allowing X-ray crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles ...
and determination of their atomic structure in three dimensions. This demonstrated that the LSm proteins share a similar three-dimensional fold of a short alpha helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand- helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues ...
and a five-stranded folded beta sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a ge ...
, subsequently named the LSm fold. Other investigations found that LSm proteins assemble into a torus
In geometry, a torus (plural tori, colloquially donut or doughnut) is a surface of revolution generated by revolving a circle in three-dimensional space about an axis that is coplanar with the circle.
If the axis of revolution does n ...
(doughnut-shaped ring) of six or seven LSm proteins, and that RNA binds to the inside of the torus, with one nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecu ...
bound to each LSm protein.
Structure
uracil
Uracil () (symbol U or Ura) is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced b ...
is stacked between the histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the ...
and arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) and both the am ...
residues, stabilized by hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
ing to an asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
residue, and hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
ing between the aspartate
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. Like all other amino acids, it contains an amino group and a carboxylic acid. Its α-amino group is in the pro ...
residue and the ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this com ...
. LSm proteins are characterized by a beta sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a ge ...
(the secondary structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary struct ...
), folded into the LSm fold (the tertiary structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may i ...
), polymerization into a six or seven member torus
In geometry, a torus (plural tori, colloquially donut or doughnut) is a surface of revolution generated by revolving a circle in three-dimensional space about an axis that is coplanar with the circle.
If the axis of revolution does n ...
(the quaternary structure), and binding to RNA oligonucleotides. A modern paradigm classifies proteins on the basis of protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monom ...
and is a currently active field, with three major approaches, SCOP (Structural Classification of Proteins), CATH
The CATH Protein Structure Classification database is a free, publicly available online resource that provides information on the evolutionary relationships of protein domains. It was created in the mid-1990s by Professor Christine Orengo and coll ...
(Class, Architecture, Topology, Homologous superfamily), and FSSP/DALI (Families of Structurally Similar Proteins).
Secondary
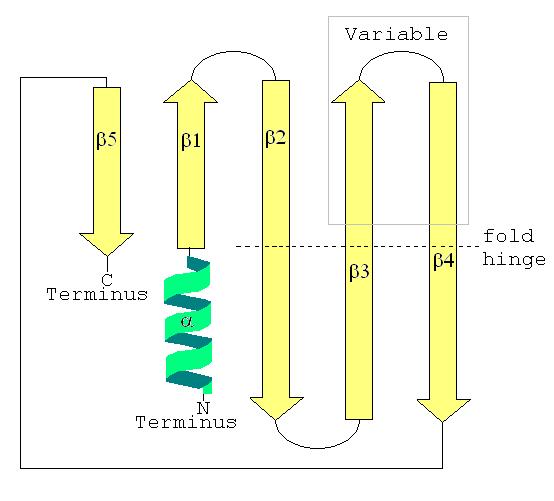
Thesecondary structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary struct ...
of a LSm protein is a small five-strand anti-parallel beta sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a ge ...
, with the strands identified from the N-terminal end
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the am ...
to the C-terminal end
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
as β1, β2, β3, β4, β5. The SCOP class of All beta proteins and the CATH class of Mainly Beta are defined as protein structures that are primarily beta sheets, thus including LSm. The SM1 sequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ' ...
corresponds to the β1, β2, β3 strands, and the SM2 sequence motif corresponds to the β4 and β5 strands. The first four beta strands are adjacent to each other, but β5 is adjacent to β1, turning the overall structure into a short barrel. This structural topology is described as 51234. A short (two to four turns) N-terminal alpha helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand- helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues ...
is also present in most LSm proteins. The β3 and β4 strands are short in some LSm proteins, and are separated by an unstructured coil of variable length. The β2, β3 and β4 strands are strongly bent about 120° degrees at their midpoints The bends in these strands are often glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid ( carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinog ...
, and the side chains internal to the beta barrel are often the hydrophobic residues valine
Valine (symbol Val or V) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- carboxylic acid group (which is in the deprotona ...
, leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- ...
, isoleucine
Isoleucine (symbol Ile or I) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depr ...
and methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical ...
.
Tertiary
SCOP simply classifies the LSm structure as the Sm-like fold, one of 149 different Beta Protein folds, without any intermediate groupings. The LSm beta sheet is sharply bent and described as a Roll architecture in CATH (one of 20 different beta protein architectures in CATH). One of the beta strands (β5 in LSm) crosses the open edge of the roll to form a small SH3 type barrel topology (one of 33 beta roll topologies in CATH). CATH lists 23 homologous superfamilies with an SH3 type barrel topology, one of which is the LSm structure (RNA Binding Protein in the CATH system). SCOP continues its structural classification after Fold to Superfamily, Family and Domain, while CATH continues to Sequence Family, but these divisions are more appropriately described in the "Evolution and phylogeny" section. The SH3-type barreltertiary structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may i ...
of the LSm fold is formed by the strongly bent (about 120°) β2, β3 and β4 strands, with the barrel structure closed by the β5 strand. Emphasizing the tertiary structure, each bent beta strand can be described as two shorter beta strands. The LSm fold can be viewed as an eight-strand anti-parallel beta sandwich, with five strands in one plane and three strands in a parallel plane with about a 45° pitch angle between the two halves of the beta sandwich. The short (two to four turns) N-terminal alpha helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand- helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues ...
occurs at one edge of the beta sandwich. This alpha helix and the beta strands can be labeled (from the N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
to the C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein i ...
) α, β1, β2a, β2b, β3a, β3b, β4a, β4b, β5 where the a and b refer to either the two halves of a bent strand in the five-strand description, or to the individual strands in the eight-strand description. Each strand (in the eight-strand description) is formed from five amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
residues. Including the bends and loops between the strands, and the alpha helix, about 60 amino acid residues contribute to the LSm fold, but this varies between homologs due to variation in inter-strand loops, the alpha helix, and even the lengths of β3b and β4a strands.
Quaternary
LSm proteins typically assemble into a LSm ring, a six or seven membertorus
In geometry, a torus (plural tori, colloquially donut or doughnut) is a surface of revolution generated by revolving a circle in three-dimensional space about an axis that is coplanar with the circle.
If the axis of revolution does n ...
, about 7 nanometers
330px, Different lengths as in respect to the molecular scale.
The nanometre (international spelling as used by the International Bureau of Weights and Measures; SI symbol: nm) or nanometer (American and British English spelling differences#-re ...
in diameter with a 2 nanometer hole. The ancestral condition is a homohexamer or homoheptamer of identical LSm subunits. LSm proteins in eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
s form heteroheptamers of seven different LSm subunits, such as the Sm proteins. Binding between the LSm proteins is best understood with the eight-strand description of the LSm fold. The five-strand half of the beta sandwich of one subunit aligns with the three-strand half of the beta sandwich of the adjacent subunit, forming a twisted 8-strand beta sheet Aβ4a/Aβ3b/Aβ2a/Aβ1/Aβ5/Bβ4b/Bβ3a/Bβ2b, where the A and B refer to the two different subunits. In addition to hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
ing between the Aβ5 and Bβ4b beta strands of the two LSm protein subunits, there are energetically favorable contacts between hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water.
Hydrophobic molecules tend to be nonpolar and, ...
amino acid side chains in the interior of the contact area, and energetically favorable contacts between hydrophilic
A hydrophile is a molecule or other molecular entity that is attracted to water molecules and tends to be dissolved by water.Liddell, H.G. & Scott, R. (1940). ''A Greek-English Lexicon'' Oxford: Clarendon Press.
In contrast, hydrophobes are n ...
amino acid side chains around the periphery of the contact area.
RNA oligonucleotide binding
LSm rings form ribonucleoprotein complexes with RNAoligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
s that vary in binding strength from very stable complexes (such as the Sm class snRNPs) to transient complexes. RNA oligonucleotides generally bind inside the hole (lumen) of the LSm torus, one nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecu ...
per LSm subunit, but additional nucleotide binding sites have been reported at the top (α helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues ea ...
side) of the ring. The exact chemical nature of this binding varies, but common motifs include stacking the heterocyclic base (often uracil
Uracil () (symbol U or Ura) is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced b ...
) between planar side chains of two amino acids, hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
ing to the heterocyclic base and/or the ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this com ...
, and salt bridges to the phosphate
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid .
The phosphate or orthophosphate ion is derived from phosph ...
group.
Functions
The various kinds of LSm rings function as scaffolds or chaperones for RNAoligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
s, assisting the RNA to assume and maintain the proper three-dimensional structure. In some cases, this allows the oligonucleotide RNA to function catalytically as a ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demons ...
. In other cases, this facilitates modification or degradation of the RNA, or the assembly, storage, and intracellular transport of ribonucleoprotein complexes.
Sm ring
The Sm ring is found in thenucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
of all eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
s (about 2.5 x 106 copies per proliferating human cell), and has the best understood functions. The Sm ring is a heteroheptamer. The Sm-class snRNA molecule (in the 5' to 3' direction) enters the lumen (doughnut hole) at the SmE subunit and proceeds sequentially in a clockwise fashion (looking from the α helix side) inside the lumen (doughnut hole) to the SmG, SmD3, SmB, SmD1, SmD2 subunits, exiting at the SmF subunit. (SmB can be replaced by the splice variant SmB' and by SmN in neural tissues.) The Sm ring permanently binds to the U1, U2, U4 and U5 snRNAs which form four of the five snRNPs that constitute the major spliceosome. The Sm ring also permanently binds to the U11, U12 and U4atac snRNAs which form four of the five snRNPs (including the U5 snRNP) that constitute the minor spliceosome The minor spliceosome is a ribonucleoprotein complex that catalyses the removal ( splicing) of an atypical class of spliceosomal introns (U12-type) from messenger RNAs in some clades of eukaryotes. This process is called noncanonical splicing, as op ...
. Both of these spliceosomes are central RNA-processing complexes in the maturation of messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
from pre-mRNA. Sm proteins have also been reported to be part of ribonucleoprotein component of telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most euk ...
.
Lsm2-8 ring
The two Lsm2-8 snRNPs (U6 and U6atac) have the key catalytic function in the major and minor spliceosomes. These snRNPs do not include the Sm ring, but instead use the heteroheptameric Lsm2-8 ring. The LSm rings are about 20 times less abundant than the Sm rings. The order of these seven LSm proteins in this ring is not known, but based on amino acid sequence homology with the Sm proteins, it is speculated that the snRNA (in the 5' to 3' direction) may bind first to LSm5, and precedes sequentially clockwise to the LSm7, LSm4, LSm8, LSm2, LSm3, and exiting at the LSm6 subunit. Experiments with ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have b ...
'' (budding yeast) mutations suggest that the Lsm2-8 ring assists the reassociation of the U4 and U6 snRNPs into the U4/U6 di-snRNP. (After completion of exon deletion and intron splicing, these two snRNPs must reassociate for the spliceosome to initiate another exon/intron splicing cycle. In this role, the Lsm2-8 ring acts as an RNA chaperone instead of an RNA scaffold.) The Lsm2-8 ring also forms an snRNP with the U8 small nucleolar RNA
In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, t ...
(snoRNA) which localizes in the nucleolus
The nucleolus (, plural: nucleoli ) is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis, which is the synthesis of ribosomes. The nucleolus also participates in the formation of sign ...
. This ribonucleoprotein complex is necessary for processing ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from riboso ...
and transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
to their mature forms. The Lsm2-8 ring is reported to have a role in the processing of pre-P RNA into RNase P RNA. In contrast to the Sm ring, the Lsm2-8 ring does not permanently bind to its snRNA and snoRNA.
Sm10/Sm11 ring
A second type of Sm ring exists where LSm10 replaces SmD1 and LSm11 replaces SmD2. LSm11 is a two domain protein with theC-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
domain being a LSm domain. This heteroheptamer ring binds with the U7 snRNA in the U7 snRNP. The U7 snRNP mediates processing of the 3' UTR stem-loop of the histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
in the nucleus. Like the Sm ring, it is assembled in the cytoplasm onto the U7 snRNA by a specialized SMN complex.
Lsm1-7 ring
A second type of Lsm ring is the Lsm1-7 ring, which has the same structure as the Lsm2-8 ring except that LSm1 replaces LSm8. In contrast to the Lsm2-8 ring, the Lsm1-7 ring localizes in thecytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
where it assists in degrading messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
in ribonucleoprotein complexes. This process controls the turnover of messenger RNA so that ribosomal translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
of mRNA to protein responds quickly to changes in transcription of DNA to messenger RNA by the cell. LSM1-7, together with Pat1, has been shown to play a role in the formation of P-bodies after deadenylation.
Gemin6 and Gemin7
The SMN complex (described under "Biogenesis of snRNPs") is composed of the SMN protein and Gemin2-8. Two of these, Gemin 6 and Gemin7 have been discovered to have the LSm structure, and to form a heterodimer. These may have a chaperone function in the SMN complex to assist the formation of the Sm ring on the Sm-class snRNAs. PRMT5 complex is composed ofPRMT5
Protein arginine N-methyltransferase 5 is an enzyme that in humans is encoded by the ''PRMT5'' gene. PRMT5 symmetrically dimethylates H2AR3, H4R3, H3R2, and H3R8 in vivo, all of which are linked to a range of transcriptional regulatory events.
PR ...
, pICln, WD45 (Mep50). pICln helps to form Sm opened ring on SMN complex. SMN complex assists in the assembly of snRNPs where the Sm ring is in the open conformation on SMN complex and this Sm ring is loaded onto the snRNA by SMN complex.
LSm12-16 and other multi-domain LSm proteins
The LSm12-16 proteins have been described very recently. These are two-domain proteins with aN-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
LSm domain and a C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
methyl transferase domain. Very little is known about the function of these proteins, but presumably they are member of LSm-domain rings that interact with RNA. There is some evidence that LSm12 is possibly involved in mRNA degradation and LSm13-16 may have roles in regulation of mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintai ...
. Unexpectedly, LSm12 was recently implicated in Calcium signaling by acting as the intermediate binding-protein for the nucleotide second messenger, NAADP ( Nicotinic acid adenine dinucleotide phosphate) that activates endo-lysosomal Ca2+ channels TPCs ( Two-pore channels). This occurred by NAADP binding to the LSm domain, not the AD domain. A large protein of unknown function, ataxin-2, associated with the neurodegenerative disease spinocerebellar ataxia type 2, also has a N-terminal LSm domain.
Archaeal Sm rings
Two LSm proteins are found in a seconddomain
Domain may refer to:
Mathematics
*Domain of a function, the set of input values for which the (total) function is defined
** Domain of definition of a partial function
** Natural domain of a partial function
**Domain of holomorphy of a function
* ...
of life, the Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaeba ...
. These are the Sm1 and Sm2 proteins (not to be confused with the Sm1 and Sm2 sequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ' ...
s), and are sometimes identified as Sm-like archaeal proteins SmAP1 and SmAP2 for this reason. Sm1 and Sm2 generally form homoheptamer rings, although homohexamer rings have been observed. Sm1 rings are similar to eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
Lsm rings in that they form in the absence of RNA while Sm2 rings are similar to eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
Sm rings in that they require uridine-rich RNA for their formation. They have been reported to associate with RNase P RNA, suggesting a role in transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
processing, but their function in archaea in this process (and possibly processing other RNA such as ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from riboso ...
) is mostly unknown. One of the two main branches of archaea, the crenarchaeotes have a third known type of archaeal LSm protein, Sm3. This is a two-domain protein with a N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
LSm domain that forms a homoheptamer ring. Nothing is known about the function of this LSm protein, but presumably it interacts with, and probably helps process, RNA in these organisms.
Bacterial LSm rings
Several LSm proteins have been reported in the thirddomain
Domain may refer to:
Mathematics
*Domain of a function, the set of input values for which the (total) function is defined
** Domain of definition of a partial function
** Natural domain of a partial function
**Domain of holomorphy of a function
* ...
of life, the Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
. Hfq protein forms homohexamer rings, and was originally discovered as necessary for infection by the bacteriophage Qβ
Bacteriophage Qbeta (''Qubevirus durum''), commonly referred to as Qbeta or Qβ, is a positive-strand RNA virus which infects bacteria that have F-pili, most commonly ''Escherichia coli''. Its linear genome is packaged into an icosahedral cap ...
, although this is clearly not the native function of this protein in bacteria. It is not universally present in all bacteria, but has been found in Pseudomonadota
Pseudomonadota (synonym Proteobacteria) is a major phylum of Gram-negative bacteria. The renaming of phyla in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature. Th ...
, Bacillota, Spirochaetota, Thermotogota, Aquificota, and one species of Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaeba ...
. (This last instance is probably a case of horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). H ...
.) Hfq is pleiotropic
Pleiotropy (from Greek , 'more', and , 'way') occurs when one gene influences two or more seemingly unrelated phenotypic traits. Such a gene that exhibits multiple phenotypic expression is called a pleiotropic gene. Mutation in a pleiotropic ge ...
with a variety of interactions, generally associated with translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
regulation. These include blocking ribosome binding to mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
, marking mRNA for degradation by binding to their poly-A tails, and association with bacterial small regulatory RNAs (such as DsrA RNA) that control translation by binding to certain mRNAs
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
. A second bacterial LSm protein is YlxS (sometimes also called YhbC), which was first identified in the soil bacterium ''Bacillus subtilis
''Bacillus subtilis'', known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants, humans and marine sponges. As a member of the genus ''Bacillus ...
''. This is a two-domain protein with a N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
LSm domain. Its function is unknown, but amino acid sequence homologs are found in virtually every bacterial genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
to date, and it may be an essential protein. The middle domain of the small conductance mechanosensitive channel MscS in ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' forms a homoheptameric ring. This LSm domain has no apparent RNA-binding function, but the homoheptameric torus is part of the central channel of this membrane protein.
Evolution and phylogeny
LSm homologs are found in all three domains of life, and may even be found in every singleorganism
In biology, an organism () is any living system that functions as an individual entity. All organisms are composed of cells ( cell theory). Organisms are classified by taxonomy into groups such as multicellular animals, plants, and fu ...
. Computational phylogenetic
Computational phylogenetics is the application of computational algorithms, methods, and programs to phylogenetic analyses. The goal is to assemble a phylogenetic tree representing a hypothesis about the evolutionary ancestry of a set of genes, spe ...
methods are used to infer phylogenetic
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups ...
relations. Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Al ...
between the various LSm homologs are the appropriate tool for this, such as multiple sequence alignment
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutio ...
of the primary structure (amino acid sequence), and structural alignment
Structural alignment attempts to establish homology between two or more polymer structures based on their shape and three-dimensional conformation. This process is usually applied to protein tertiary structures but can also be used for large R ...
of the tertiary structure (three-dimensional structure). It is hypothesized that a gene for a LSm protein was present in the last universal ancestor of all life. Based on the functions of known LSm proteins, this original LSm protein may have assisted ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demons ...
s in the processing of RNA for synthesizing proteins as part of the RNA world hypothesis
The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existenc ...
of early life. According to this view, this gene was passed from ancestor to descendant, with frequent mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
s, gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. ...
s and occasional horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). H ...
s. In principle, this process can be summarized in a phylogenetic tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
with the root in the last universal ancestor (or earlier), and with the tips representing the universe of LSm genes existing today.
Homomeric LSm rings in bacteria and archaea
Based on structure, the known LSm proteins divide into a group consisting of the bacterial LSm proteins (Hfq, YlxS and MscS) and a second group of all other LSm proteins, in accordance with the most recently publishedphylogenetic tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
s. The three archaeal LSm proteins (Sm1, Sm2 and Sm3) also cluster as a group, distinct from the eukaryote LSm proteins. Both the bacterial and archaeal LSm proteins polymerize to homomeric rings, which is the ancestral condition.
Heteromeric LSm rings in eukaryotes
A series of gene duplications of a single eukaryote LSm gene resulted in most (if not all) of the known eukaryote LSm genes. Each of the seven Sm proteins has greater amino acid sequence homology to a corresponding Lsm protein than to the other Sm proteins. This suggests that an ancestral LSm gene duplicated several times, resulting in seven paralogs. These subsequently diverged from each other so that the ancestral homoheptamer LSm ring became a heteroheptamer ring. Based on the known functions of LSm proteins in eukaryotes and archaea, the ancestral function may have been processing of pre-ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from riboso ...
, pre-transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
, and pre- RNase P. Then, according to this hypothesis, the seven ancestral eukaryote LSm genes duplicated again to seven pairs of Sm/LSm paralogs; LSm1/SmB, LSm2/SmD1, LSm3/SmD2, LSm4/SmD3, LSm5/SmE, LSm6/SmF and LSm7/SmG. These two group of seven LSm genes (and the corresponding two kinds of LSm rings) evolved to an Sm ring (requiring RNA) and a Lsm ring (which forms without RNA). The LSm1/LSm8 paralog pair also seems to have originated prior to the last common eukaryote ancestor, for a total of at least 15 LSm protein genes. The SmD1/LSm10 paralog pair and the SmD2/LSm11 paralog pair exist only in animal
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia. With few exceptions, animals consume organic material, breathe oxygen, are able to move, can reproduce sexually, and go through an ontogenetic stage ...
s, fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
, and the amoebozoa
Amoebozoa is a major taxonomic group containing about 2,400 described species of amoeboid protists, often possessing blunt, fingerlike, lobose pseudopods and tubular mitochondrial cristae. In traditional and currently no longer supported c ...
(sometimes identified as the unikont clade) and appears to be absent in the bikont clade (chromalveolate
Chromalveolata was a eukaryote supergroup present in a major classification of 2005, then regarded as one of the six major groups within the eukaryotes.
It was a refinement of the kingdom Chromista, first proposed by Thomas Cavalier-Smith in ...
s, excavates, plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s and rhizaria
The Rhizaria are an ill-defined but species-rich supergroup of mostly unicellular eukaryotes. Except for the Chlorarachniophytes and three species in the genus Paulinella in the phylum Cercozoa, they are all non-photosynthethic, but many forami ...
). Therefore, these two gene duplications predated this fundamental split in the eukaryote lineage. The SmB/SmN paralog pair is seen only in the placental mammals, which dates this LSm gene duplication.
Biogenesis of snRNPs
Small nuclear ribonucleoproteins (snRNPs) assemble in a tightly orchestrated and regulated process that involves both thecell nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, h ...
and cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
.
References
External links
Pfam entry LSM. Pfam is the Sanger Institute database, which is a collection of protein families and domains.
{{DEFAULTSORT:Lsm Protein families Spliceosome