Biomethylation on:
[Wikipedia]
[Google]
[Amazon]
In the chemical sciences, methylation denotes the addition of a
 In reverse methanogenesis, methane serves as the methylating agent.
In reverse methanogenesis, methane serves as the methylating agent.
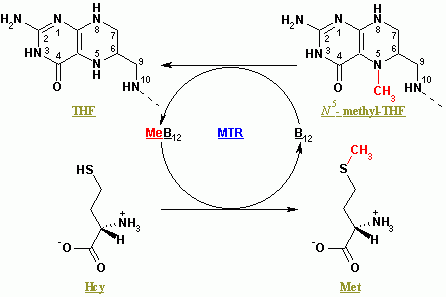 Methionine synthase regenerates methionine (Met) from homocysteine (Hcy). The overall reaction transforms 5-methyltetrahydrofolate (N5-MeTHF) into tetrahydrofolate (THF) while transferring a methyl group to Hcy to form Met. Methionine Synthases can be cobalamin-dependent and cobalamin-independent: Plants have both, animals depend on the methylcobalamin-dependent form.
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by two steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N5-MeTHF to Co(I) in enzyme-bound cobalamin (Cob), forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. Then, a Hcy that has coordinated to an enzyme-bound zinc to form a reactive thiolate reacts with the Me-Cob. The activated methyl group is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.
Methionine synthase regenerates methionine (Met) from homocysteine (Hcy). The overall reaction transforms 5-methyltetrahydrofolate (N5-MeTHF) into tetrahydrofolate (THF) while transferring a methyl group to Hcy to form Met. Methionine Synthases can be cobalamin-dependent and cobalamin-independent: Plants have both, animals depend on the methylcobalamin-dependent form.
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by two steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N5-MeTHF to Co(I) in enzyme-bound cobalamin (Cob), forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. Then, a Hcy that has coordinated to an enzyme-bound zinc to form a reactive thiolate reacts with the Me-Cob. The activated methyl group is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.
RCO2H + tmsCHN2 + CH3OH -> RCO2CH3 + CH3Otms + N2
The method offers the advantage that the side products are easily removed from the product mixture.
deltaMasses
Detection of Methylations after Mass Spectrometry {{Authority control Methylation, Epigenetics Organic reactions Post-translational modification
methyl group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula . In formulas, the group is often abbreviated as Me. This hydrocarbon group occurs in ma ...
on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation
Alkylation is the transfer of an alkyl group from one molecule to another. The alkyl group may be transferred as an alkyl carbocation, a free radical, a carbanion, or a carbene (or their equivalents). Alkylating agents are reagents for effecti ...
, with a methyl group replacing a hydrogen
Hydrogen is the chemical element with the symbol H and atomic number 1. Hydrogen is the lightest element. At standard conditions hydrogen is a gas of diatomic molecules having the formula . It is colorless, odorless, tasteless, non-toxic ...
atom. These terms are commonly used in chemistry, biochemistry
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and ...
, soil science
Soil science is the study of soil as a natural resource on the surface of the Earth including soil formation, classification and mapping; physical, chemical, biological, and fertility properties of soils; and these properties in relation to th ...
, and the biological sciences
Biology is the scientific study of life. It is a natural science with a broad scope but has several unifying themes that tie it together as a single, coherent field. For instance, all organisms are made up of cells that process hereditary ...
.
In biological systems, methylation is catalyzed
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of Protein#Functions, protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain Histology#Histological Artifacts, histological staining artifacts. The reverse of methylation is demethylation.
In biology
In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics.Methanogenesis
Methanogenesis, the process that generates methane from CO2, involves a series of methylation reactions. These reactions are effected by a set of enzymes harbored by a family of anaerobic microbes. In reverse methanogenesis, methane serves as the methylating agent.
In reverse methanogenesis, methane serves as the methylating agent.
O-methyltransferases
A wide variety of phenols undergo O-methylation to give anisole derivatives. This process, catalyzed by enzymes such as caffeoyl-CoA O-methyltransferase, is a key reaction in the biosynthesis of Monolignol, lignols, precursor (chemistry), percursors to lignin, a major structural component of plants. Plants produce flavonoids and isoflavones with methylations on hydroxyl groups, i.e. methoxy group, methoxy bonds. This 5-O-methylation affects the flavonoid´s water solubility. Examples are 5-O-Methylgenistein, 5-O-methylgenistein, 5-O-Methylmyricetin, 5-O-methylmyricetin or 5-O-Methylquercetin, 5-O-methylquercetin, also known as azaleatin.Proteins
Together with ubiquitin and phosphorylation, methylation is a major biochemical process for modifying protein function. The most prevalent protein methylations affect arginine and lysine residue of specific histones. Otherwise histidine, glutamate, asparagine, cysteine are susceptible to methylation. Some of these products include S-Methylcysteine, ''S''-methylcysteine, two isomers of ''N''-methylhistidine, and two isomers of ''N''-methylarginine.Methionine synthase
 Methionine synthase regenerates methionine (Met) from homocysteine (Hcy). The overall reaction transforms 5-methyltetrahydrofolate (N5-MeTHF) into tetrahydrofolate (THF) while transferring a methyl group to Hcy to form Met. Methionine Synthases can be cobalamin-dependent and cobalamin-independent: Plants have both, animals depend on the methylcobalamin-dependent form.
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by two steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N5-MeTHF to Co(I) in enzyme-bound cobalamin (Cob), forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. Then, a Hcy that has coordinated to an enzyme-bound zinc to form a reactive thiolate reacts with the Me-Cob. The activated methyl group is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.
Methionine synthase regenerates methionine (Met) from homocysteine (Hcy). The overall reaction transforms 5-methyltetrahydrofolate (N5-MeTHF) into tetrahydrofolate (THF) while transferring a methyl group to Hcy to form Met. Methionine Synthases can be cobalamin-dependent and cobalamin-independent: Plants have both, animals depend on the methylcobalamin-dependent form.
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by two steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N5-MeTHF to Co(I) in enzyme-bound cobalamin (Cob), forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. Then, a Hcy that has coordinated to an enzyme-bound zinc to form a reactive thiolate reacts with the Me-Cob. The activated methyl group is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.
Heavy metals: arsenic, mercury, cadmium
Biomethylation is the pathway for converting some heavy elements into more mobile or more lethal derivatives that can enter the food chain. The biomethylation of arsenic compounds starts with the formation of methanearsonates. Thus, trivalent inorganic arsenic compounds are methylated to give methanearsonate. S-adenosylmethionine is the methyl donor. The methanearsonates are the precursors to dimethylarsonates, again by the cycle of Redox, reduction (to methylarsonous acid) followed by a second methylation. Related pathways apply to the biosynthesis of methylmercury.Epigenetic methylation
DNA/RNA methylation
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sitesthat is, where a cytosine is directly followed by a guanine in the DNA sequence). This methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG iscatalyzed
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
by the enzyme DNA methyltransferase. In mammals, DNA methylation is common in body cells, and methylation of CpG sites seems to be the default. Human DNA has about 80–90% of CpG sites methylated, but there are certain areas, known as CpG site#CpG islands, CpG islands, that are CG-rich (high cytosine and guanine content, made up of about 65% CG Residue (chemistry), residues), wherein none is methylated. These are associated with the Promoter (genetics), promoters of 56% of mammalian genes, including all Housekeeping gene, ubiquitously expressed genes. One to two percent of the human genome are CpG clusters, and there is an inverse relationship between CpG methylation and transcriptional activity. Methylation contributing to epigenetic inheritance can occur through either DNA methylation or protein methylation. Improper methylations of human genes can lead to disease development, including cancer.
Similarly, RNA methylation occurs in different RNA species viz. tRNA, rRNA, mRNA, tmRNA, snRNA, snoRNA, miRNA, and viral RNA. Different catalytic strategies are employed for RNA methylation by a variety of RNA-methyltransferases. RNA methylation is thought to have existed before DNA methylation in the early forms of life evolving on earth.
N6-Methyladenosine, N6-methyladenosine (m6A) is the most common and abundant methylation modification in RNA molecules (mRNA) present in eukaryotes. 5-methylcytosine (5-mC) also commonly occurs in various RNA molecules. Recent data strongly suggest that m6A and 5-mC RNA methylation affects the regulation of various biological processes such as RNA stability and mRNA translation, and that abnormal RNA methylation contributes to etiology of human diseases.
Protein methylation
Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence. Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylarginine) or one on both nitrogens (symmetric dimethylarginine), by protein arginine methyltransferases (PRMTs). Lysine can be methylated once, twice, or three times by lysine methyltransferases. Protein methylation has been most studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones that are methylated on certain residues can act Epigenetics, epigenetically to repress or activate gene expression. Protein methylation is one type of post-translational modification.Evolution
Methyl metabolism is very ancient and can be found in all organisms on earth, from bacteria to humans, indicating the importance of methyl metabolism for physiology. Indeed, pharmacological inhibition of global methylation in species ranging from human, mouse, fish, fly, round worm, plant, algae and cyanobacteria causes the same effects on their biological rhythms, demonstrating conserved physiological roles of methylation during evolution.In chemistry
The term methylation in organic chemistry refers to thealkylation
Alkylation is the transfer of an alkyl group from one molecule to another. The alkyl group may be transferred as an alkyl carbocation, a free radical, a carbanion, or a carbene (or their equivalents). Alkylating agents are reagents for effecti ...
process used to describe the delivery of a group.
Electrophilic methylation
Methylations are commonly performed using electrophile, ''electrophilic'' methyl sources such as iodomethane, dimethyl sulfate, dimethyl carbonate, or tetramethylammonium chloride. Less common but more powerful (and more dangerous) methylating reagents include methyl triflate, diazomethane, and methyl fluorosulfonate (magic methyl). These reagents all react via SN2 nucleophilic substitutions. For example, a carboxylate may be methylated on oxygen to give a methyl ester; an alkoxide salt may be likewise methylated to give an ether, ; or a ketone enolate may be methylated on carbon to produce a new ketone. : The Purdie methylation is a specific for the methylation at oxygen of carbohydrates using iodomethane and silver oxide. :Eschweiler–Clarke methylation
The Eschweiler–Clarke reaction is a method for methylation of amines. This method avoids the risk of quaternization, which occurs when amines are methylated with methyl halides.Diazomethane and trimethylsilyldiazomethane
Diazomethane and the safer analogue trimethylsilyldiazomethane methylate carboxylic acids, phenols, and even alcohols: :Nucleophilic methylation
Methylation sometimes involve use of nucleophile, ''nucleophilic'' methyl reagents. Strongly nucleophilic methylating agents include methyllithium () or Grignard reagents such as methylmagnesium bromide (). For example, will add methyl groups to the carbonyl (C=O) of ketones and aldehyde.: : Milder methylating agents include tetramethyltin, dimethylzinc, and trimethylaluminium.See also
Biology topics
*Bisulfite sequencing – the biochemical method used to determine the presence or absence of methyl groups on a DNA sequence *MethDB DNA Methylation Database *Microscale thermophoresis – a biophysical method to determine the methylisation state of DNAOrganic chemistry topics
*Alkylation *Methoxy *Organozinc compound#Titanium–zinc methylenation, Titanium–zinc methylenation *Petasis reagent *Nysted reagent *Wittig reaction *Tebbe's reagentReferences
External links
deltaMasses
Detection of Methylations after Mass Spectrometry {{Authority control Methylation, Epigenetics Organic reactions Post-translational modification