|
VNTR
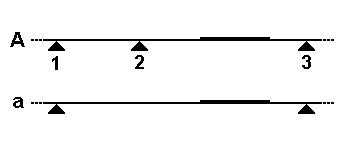
A variable number tandem repeat (or VNTR) is a location in a genome where a short nucleotide sequence is organized as a tandem repeat. These can be found on many chromosomes, and often show variations in length (number of repeats) among individuals. Each variant acts as an inherited allele, allowing them to be used for personal or parental identification. Their analysis is useful in genetics and biology research, forensics, and DNA fingerprinting. Structure and allelic variation In the schematic above, the rectangular blocks represent each of the repeated DNA sequences at a particular VNTR location. The repeats are in tandem – i.e. they are clustered together and oriented in the same direction. Individual repeats can be removed from (or added to) the VNTR via recombination or replication errors, leading to alleles with different numbers of repeats. Flanking regions are segments of repetitive sequence (shown here as thin lines), allowing the VNTR blocks to be extracted ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Restriction Fragment Length Polymorphism
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, populations, or species or to pinpoint the locations of genes within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of restriction enzyme sites, or to a related laboratory technique by which such differences can be illustrated. In RFLP analysis, a DNA sample is digested into fragments by one or more restriction enzymes, and the resulting ''restriction fragments'' are then separated by gel electrophoresis according to their size. RFLP analysis is now largely obsolete due to the emergence of inexpensive DNA sequencing technologies, but it was the first DNA profiling technique inexpensive enough to see widespread application. RFLP analysis was an important early tool in genome mapping, localization of genes for genetic disorders, determination of risk for disease, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Tandem Repeat
In genetics, tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other, e.g. ATTCG ATTCG ATTCG, in which the sequence ATTCG is repeated three times. Several protein domains also form tandem repeats within their amino acid primary structure, such as armadillo repeats. However, in proteins, perfect tandem repeats are rare in naturally proteins, but they have been added to designed proteins. Tandem repeats constitute about 8% of the human genome. They are implicated in more than 50 lethal human diseases, including amyotrophic lateral sclerosis, Huntington's disease, and several cancers. Terminology All tandem repeat arrays are classifiable as satellite DNA, a name originating from the fact that tandem DNA repeats, by nature of repeating the same nucleotide sequences repeatedly, have a unique ratio of the two possible nucleotide base pair combinations, conferring them a specific mass density that a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
DNA Fingerprinting
DNA profiling (also called DNA fingerprinting and genetic fingerprinting) is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding. DNA profiling is a forensic technique in criminal investigations, comparing criminal suspects' profiles to DNA evidence so as to assess the likelihood of their involvement in the crime. It is also used in paternity testing, to establish immigration eligibility, and in genealogical and medical research. DNA profiling has also been used in the study of animal and plant populations in the fields of zoology, botany, and agriculture. Background Starting in the mid 1970s, scientific advances allowed the use of DNA as a material for the identification of an individual. The first patent covering the direct use of DNA variation for forensicsUS5593832A was filed by Jeffrey Glassberg in 1983, based upon work he had done while ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Genetic Marker
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed. A genetic marker may be a short DNA sequence, such as a sequence surrounding a single base-pair change ( single nucleotide polymorphism, SNP), or a long one, like minisatellites. Background For many years, gene mapping was limited to identifying organisms by traditional phenotypes markers. This included genes that encoded easily observable characteristics, such as blood types or seed shapes. The insufficient number of these types of characteristics in several organisms limited the possible mapping efforts. This prompted the development of gene markers, which could identify genetic characteristics that are not readily observable in organisms (such as protein variation). Types Some commonly used types of genetic markers ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Genetic Linkage
Genetic linkage is the tendency of Nucleic acid sequence, DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two Genetic marker, genetic markers that are physically near to each other are unlikely to be separated onto different Chromatid, chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two Gene, genes are on a chromosome, the lower the chance of Genetic recombination, recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a physical and a functional standpoint. It started in 1990 and was completed in 2003. It was the world's largest collaborative biological project. Planning for the project began in 1984 by the US government, and it officially launched in 1990. It was declared complete on 14 April 2003, and included about 92% of the genome. Level "complete genome" was achieved in May 2021, with only 0.3% of the bases covered by potential issues. The final gapless assembly was finished in January 2022. Funding came from the US government through the National Institutes of Health (NIH) as well as numerous other groups from around the world. A parallel project was conducted outside the government by the Celera Corporation, or Celera Genomics, which was formall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Forensic
Forensic science combines principles of law and science to investigate criminal activity. Through crime scene investigations and laboratory analysis, forensic scientists are able to link suspects to evidence. An example is determining the time and cause of death through autopsies. This evidence can then be used for proof towards a crime. Forensic science, often confused with criminalistics, is the application of science principles and methods to support legal decision-making in matters of criminal and civil law. During criminal investigation in particular, it is governed by the legal standards of admissible evidence and criminal procedure. It is a broad field utilizing numerous practices such as the analysis of DNA, fingerprints, bloodstain patterns, firearms, ballistics, toxicology, microscopy, and fire debris analysis. Forensic scientists collect, preserve, and analyze evidence during the course of an investigation. While some forensic scientists travel to the scene of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Southern Blot
Southern blot is a method used for detection and quantification of a specific DNA sequence in DNA samples. This method is used in molecular biology. Briefly, purified DNA from a biological sample (such as blood or tissue) is digested with restriction enzymes, and the resulting DNA fragments are separated by electrophoresis using an electric current to move them through a sieve-like gel or matrix, which allows smaller fragments to move faster than larger fragments. The DNA fragments are transferred out of the gel or matrix onto a solid membrane, which is then exposed to a DNA probe labeled with a radioactive, fluorescent, or chemical tag. The tag allows any DNA fragments containing complementary sequences with the DNA probe sequence to be visualized within the Southern blot. The Southern blotting combines the transfer of electrophoresis-separated DNA fragments to a filter membrane in a process called blotting, and the subsequent fragment detection by probe hybridization. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
DNA Fingerprint
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life. The two DNA strands are known as polynucleotides as they are composed of simpler monomeric units called nucleotides. Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine guanine adenine or thymine , a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds (known as the phosphodiester linkage) between the sugar of one nucleotide and the phosphate of the next, resulting in a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |