|
Structural Classification Of Proteins
The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor. Similar to CATH and Pfam databases, SCOP provides a classification of individual structural domains of proteins, rather than a classification of the entire proteins which may include a significant number of different domains. The SCOP database is freely accessible on the internet. SCOP was created in 1994 in the Centre for Protein Engineering and the La ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Classification Of Proteins Database Logo
A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as organism, biological organisms, minerals and chemical substance, chemicals. Abstract structures include data structures in computer science and musical form. Types of structure include a hierarchy (a cascade of one-to-many relationships), a Complex network, network featuring many-to-many Link (geometry), links, or a lattice (order), lattice featuring connections between components that are neighbors in space. Load-bearing Buildings, aircraft, skeletons, Ant colony, anthills, beaver dams, bridges and salt domes are all examples of Structural load, load-bearing structures. The results of construction are divided into buildings and nonbuilding structure, non-building structures, and make up the infrastructure of a human society. Built str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Fold Class
In molecular biology, protein fold classes are broad categories of protein tertiary structure topology. They describe groups of proteins that share similar amino acid and secondary structure proportions. Each class contains multiple, independent protein superfamilies (i.e. are not necessarily evolutionarily related to one another). Generally recognised classes Four large classes of protein that are generally agreed upon by the two main structure classification databases (SCOP and CATH). all-α All-α proteins are a class of structural domains in which the secondary structure is composed entirely of α-helices, with the possible exception of a few isolated β-sheets on the periphery. Common examples include the bromodomain, the globin fold and the homeodomain fold. all-β All-β proteins are a class of structural domains in which the secondary structure is composed entirely of β-sheets, with the possible exception of a few isolated α-helices on the periphery. C ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dockerin
Dockerin is a protein domain found in the cellulosome cellular structure of anaerobic bacteria. It is found on many endoglucanase enzymes. The dockerin's binding partner is the cohesin domain, located on the scaffoldin protein. This interaction between the dockerin domains of the enzyme constituents of the cellulosome and the cohesin domains of the scaffoldin protein is essential to the construction of the cellulosome complex. The Dockerin domain has two in-tandem repeats of a non-EF hand calcium binding motif. Each motif is characterized by a loop-helix structure. The three-dimensional structure of dockerin has been determined in solution,; as well as in complex with Cohesin.; There are three types of Dockerin domains: I, II and III which bind to Cohesin Type I, Cohesin Type II and Cohesin Type III respectively. A type I dockerin domain is 65-70 residues long.InterPro: The binding specificity of Type I interaction was well studied by structural and mutagenesis studies. Typ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Globin
The globins are a superfamily of heme-containing globular proteins, involved in binding and/or transporting oxygen. These proteins all incorporate the globin fold, a series of eight alpha helical segments. Two prominent members include myoglobin and hemoglobin. Both of these proteins reversibly bind oxygen via a heme prosthetic group. They are widely distributed in many organisms. Structure Globin superfamily members share a common three-dimensional fold. This 'globin fold' typically consists of eight alpha helices, although some proteins have additional helix extensions at their termini. Since the globin fold contains only helices, it is classified as an all-alpha protein fold. The globin fold is found in its namesake globin families as well as in phycocyanins. The globin fold was thus the first protein fold discovered (myoglobin was the first protein whose structure was solved). Helix packaging The eight helices of the globin fold core share significant nonlocal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coiled-coil
A coiled coil is a structural motif in proteins in which 2–7 alpha-helices are coiled together like the strands of a rope. (Dimers and trimers are the most common types.) Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-Jun, as well as the muscle protein tropomyosin. Discovery The possibility of coiled coils for α-keratin was initially somewhat controversial. Linus Pauling and Francis Crick independently came to the conclusion that this was possible at about the same time. In the summer of 1952, Pauling visited the laboratory in England where Crick worked. Pauling and Crick met and spoke about various topics; at one point, Crick asked whether Pauling had considered "coiled coils" (Crick came up with the term), to which Pauling said he had. Upon returning to the United States, Pauling resumed research on the topic. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bridges
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251&nbs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand
In coordination chemistry, a ligand is an ion or molecule ( functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electron pairs, often through Lewis bases. The nature of metal–ligand bonding can range from covalent to ionic. Furthermore, the metal–ligand bond order can range from one to three. Ligands are viewed as Lewis bases, although rare cases are known to involve Lewis acidic "ligands". Metals and metalloids are bound to ligands in almost all circumstances, although gaseous "naked" metal ions can be generated in a high vacuum. Ligands in a complex dictate the reactivity of the central atom, including ligand substitution rates, the reactivity of the ligands themselves, and redox. Ligand selection requires critical consideration in many practical areas, including bioinorganic and medicinal chemistry, homogeneous catalysis, and environment ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Protein
Small proteins are a diverse fold class of proteins (usually <100 long). Their tertiary structure is usually maintained by , metal ligands, and or such as . Some small proteins serve important regulatory functions by direct interaction with certain enzymes and are therefore also an interesting tool for biotechnological applica ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
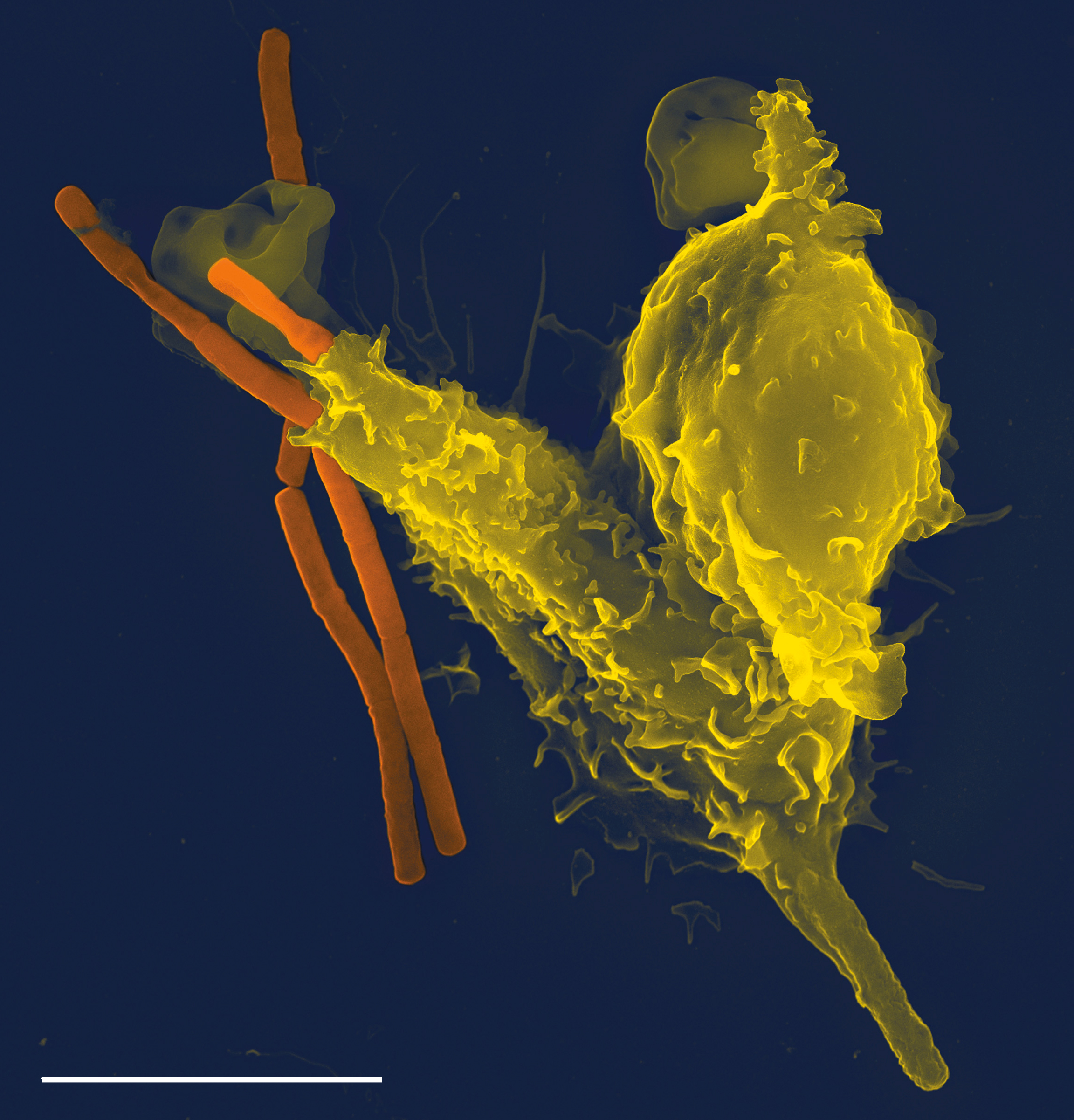
Immune System
The immune system is a network of biological processes that protects an organism from diseases. It detects and responds to a wide variety of pathogens, from viruses to parasitic worms, as well as cancer cells and objects such as wood splinters, distinguishing them from the organism's own healthy tissue. Many species have two major subsystems of the immune system. The innate immune system provides a preconfigured response to broad groups of situations and stimuli. The adaptive immune system provides a tailored response to each stimulus by learning to recognize molecules it has previously encountered. Both use molecules and cells to perform their functions. Nearly all organisms have some kind of immune system. Bacteria have a rudimentary immune system in the form of enzymes that protect against virus infections. Other basic immune mechanisms evolved in ancient plants and animals and remain in their modern descendants. These mechanisms include phagocytosis, antimicrobial ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Membrane
The cell membrane (also known as the plasma membrane (PM) or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of all cells from the outside environment (the extracellular space). The cell membrane consists of a lipid bilayer, made up of two layers of phospholipids with cholesterols (a lipid component) interspersed between them, maintaining appropriate membrane fluidity at various temperatures. The membrane also contains membrane proteins, including integral proteins that span the membrane and serve as membrane transporters, and peripheral proteins that loosely attach to the outer (peripheral) side of the cell membrane, acting as enzymes to facilitate interaction with the cell's environment. Glycolipids embedded in the outer lipid layer serve a similar purpose. The cell membrane controls the movement of substances in and out of cells and organelles, being selectively permeable to io ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


4-3D-balls.png)