Gyrase on:
[Wikipedia]
[Google]
[Amazon]
DNA gyrase, or simply gyrase, is an
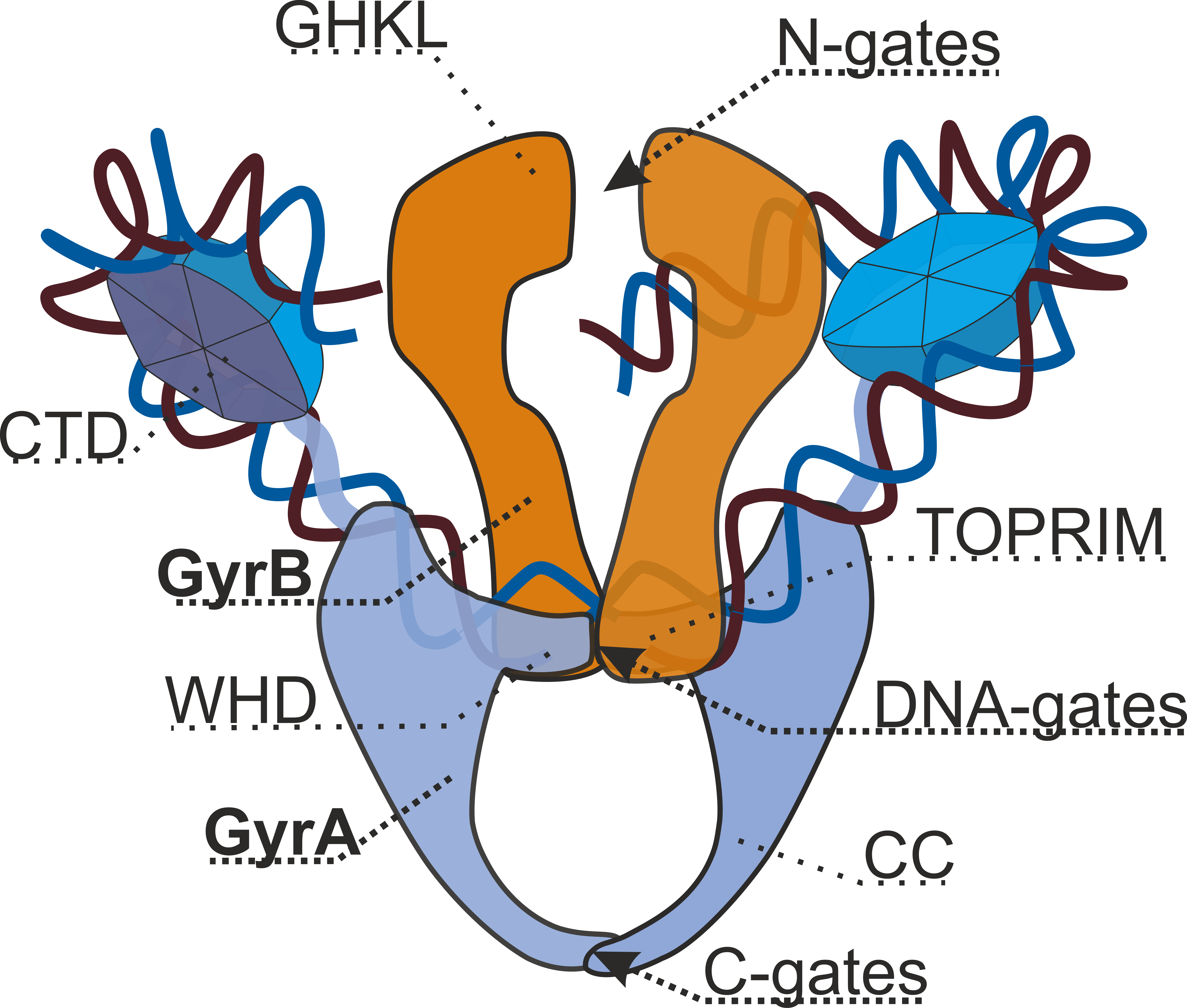 DNA gyrase is a tetrameric enzyme that consists of 2 GyrA ("A") and 2 GyrB ("B") subunits. Structurally the complex is formed by 3 pairs of "gates", sequential opening and closing of which results into the direct transfer of DNA segment and introduction of 2 negative supercoils. N-gates are formed by ATPase domains of GyrB subunits. Binding of 2 ATP molecules leads to dimerization and, therefore, closing of the gates. Hydrolysis, on the contrary, opens them. DNA cleavage and reunion is performed by a catalytic center located in DNA-gates build by all gyrase subunits. C-gates are formed by GyrA subunits.
DNA gyrase is a tetrameric enzyme that consists of 2 GyrA ("A") and 2 GyrB ("B") subunits. Structurally the complex is formed by 3 pairs of "gates", sequential opening and closing of which results into the direct transfer of DNA segment and introduction of 2 negative supercoils. N-gates are formed by ATPase domains of GyrB subunits. Binding of 2 ATP molecules leads to dimerization and, therefore, closing of the gates. Hydrolysis, on the contrary, opens them. DNA cleavage and reunion is performed by a catalytic center located in DNA-gates build by all gyrase subunits. C-gates are formed by GyrA subunits.
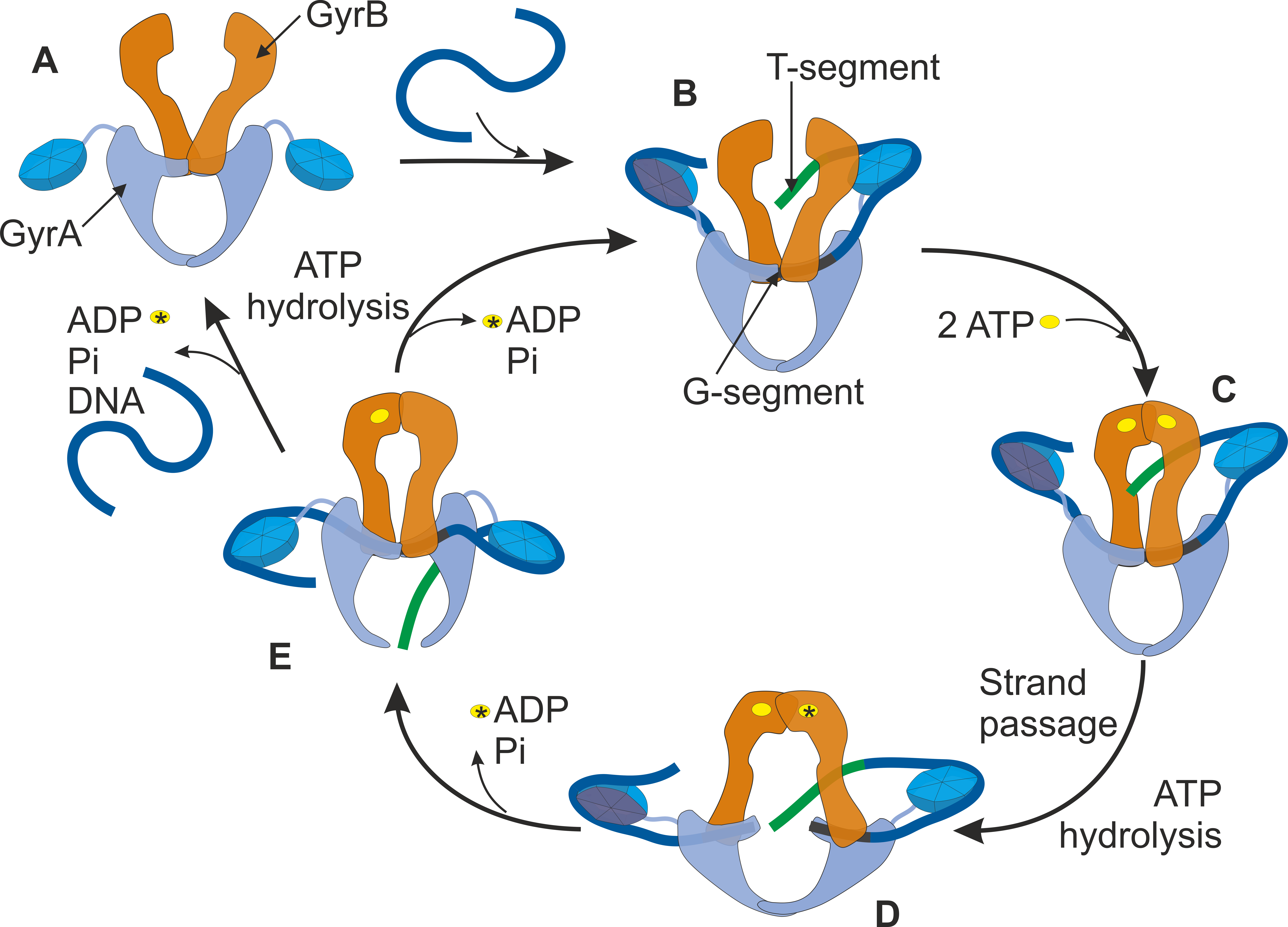 A single molecule study has characterized gyrase activity as a function of DNA tension (applied force) and ATP, and proposed a mechanochemical model. Upon binding to DNA (the "Gyrase-DNA" state), there is a competition between DNA wrapping and dissociation, where increasing DNA tension increases the probability of dissociation. According to the catalytic cycle proposed, binding of 2 ATP molecules causes dimerization of ATPase domains of GyrB subunits and capturing of a T-segment of DNA (T- from ''transferring'') in a cavity between GyrB subunits. On a next step the enzyme cleaves a G-segment of DNA (G- from ''gate'') making a
A single molecule study has characterized gyrase activity as a function of DNA tension (applied force) and ATP, and proposed a mechanochemical model. Upon binding to DNA (the "Gyrase-DNA" state), there is a competition between DNA wrapping and dissociation, where increasing DNA tension increases the probability of dissociation. According to the catalytic cycle proposed, binding of 2 ATP molecules causes dimerization of ATPase domains of GyrB subunits and capturing of a T-segment of DNA (T- from ''transferring'') in a cavity between GyrB subunits. On a next step the enzyme cleaves a G-segment of DNA (G- from ''gate'') making a
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. ...
within the class of topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues i ...
and is a subclass of Type II topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change th ...
s that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating ...
in front of the progressing replication fork
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance ...
. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number
In mathematics, the linking number is a numerical invariant that describes the linking of two closed curves in three-dimensional space. Intuitively, the linking number represents the number of times that each curve winds around the other. In E ...
by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastid
The plastid (Greek: πλαστός; plastós: formed, molded – plural plastids) is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. They are considered to be intracellular endosymbiotic cyanoba ...
s: it has been found in the apicoplast
An apicoplast is a derived non-photosynthetic plastid found in most Apicomplexa, including ''Toxoplasma gondii'', and ''Plasmodium falciparum'' and other ''Plasmodium'' spp. (parasites causing malaria), but not in others such as ''Cryptosporidium' ...
of the malarial parasite ''Plasmodium falciparum
''Plasmodium falciparum'' is a unicellular protozoan parasite of humans, and the deadliest species of ''Plasmodium'' that causes malaria in humans. The parasite is transmitted through the bite of a female ''Anopheles'' mosquito and causes the di ...
'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention ...
s, including nalidixic acid
Nalidixic acid (tradenames Nevigramon, NegGram, Wintomylon and WIN 18,320) is the first of the synthetic quinolone antibiotics.
In a technical sense, it is a naphthyridone, not a quinolone: its ring structure is a 1,8-naphthyridine nucleus that ...
, novobiocin
Novobiocin, also known as albamycin or cathomycin, is an aminocoumarin antibiotic that is produced by the actinomycete '' Streptomyces niveus'', which has recently been identified as a subjective synonym for ''S. spheroides'' a member of the clas ...
, albicidin and ciprofloxacin
Ciprofloxacin is a fluoroquinolone antibiotic used to treat a number of bacterial infections. This includes bone and joint infections, intra abdominal infections, certain types of infectious diarrhea, respiratory tract infections, skin infe ...
.
The unique ability of gyrase to introduce negative supercoils into DNA at the expense of ATP hydrolysis is what allows bacterial DNA to have free negative supercoils. The ability of gyrase to relax positive supercoils comes into play during DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance ...
and prokaryotic transcription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
. The helical nature of the DNA causes positive supercoils to accumulate ahead of a translocating enzyme, in the case of DNA replication, a DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
. The ability of gyrase (and topoisomerase IV
Topoisomerase IV is one of two Type II topoisomerases in bacteria, the other being DNA gyrase. Like gyrase, topoisomerase IV is able to pass one double-strand of DNA through another double-strand of DNA, thereby changing the linking number of DNA b ...
) to relax positive supercoils allows superhelical tension ahead of the polymerase to be released so that replication can continue.
Gyrase structure
 DNA gyrase is a tetrameric enzyme that consists of 2 GyrA ("A") and 2 GyrB ("B") subunits. Structurally the complex is formed by 3 pairs of "gates", sequential opening and closing of which results into the direct transfer of DNA segment and introduction of 2 negative supercoils. N-gates are formed by ATPase domains of GyrB subunits. Binding of 2 ATP molecules leads to dimerization and, therefore, closing of the gates. Hydrolysis, on the contrary, opens them. DNA cleavage and reunion is performed by a catalytic center located in DNA-gates build by all gyrase subunits. C-gates are formed by GyrA subunits.
DNA gyrase is a tetrameric enzyme that consists of 2 GyrA ("A") and 2 GyrB ("B") subunits. Structurally the complex is formed by 3 pairs of "gates", sequential opening and closing of which results into the direct transfer of DNA segment and introduction of 2 negative supercoils. N-gates are formed by ATPase domains of GyrB subunits. Binding of 2 ATP molecules leads to dimerization and, therefore, closing of the gates. Hydrolysis, on the contrary, opens them. DNA cleavage and reunion is performed by a catalytic center located in DNA-gates build by all gyrase subunits. C-gates are formed by GyrA subunits.
Mechanochemical model of gyrase activity
 A single molecule study has characterized gyrase activity as a function of DNA tension (applied force) and ATP, and proposed a mechanochemical model. Upon binding to DNA (the "Gyrase-DNA" state), there is a competition between DNA wrapping and dissociation, where increasing DNA tension increases the probability of dissociation. According to the catalytic cycle proposed, binding of 2 ATP molecules causes dimerization of ATPase domains of GyrB subunits and capturing of a T-segment of DNA (T- from ''transferring'') in a cavity between GyrB subunits. On a next step the enzyme cleaves a G-segment of DNA (G- from ''gate'') making a
A single molecule study has characterized gyrase activity as a function of DNA tension (applied force) and ATP, and proposed a mechanochemical model. Upon binding to DNA (the "Gyrase-DNA" state), there is a competition between DNA wrapping and dissociation, where increasing DNA tension increases the probability of dissociation. According to the catalytic cycle proposed, binding of 2 ATP molecules causes dimerization of ATPase domains of GyrB subunits and capturing of a T-segment of DNA (T- from ''transferring'') in a cavity between GyrB subunits. On a next step the enzyme cleaves a G-segment of DNA (G- from ''gate'') making a double-strand break
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dama ...
. Then the T-segment is transferred through the break, which is accompanied by the hydrolysis of the first ATP molecule. DNA-gyrase ligates the break in a G-segment back and T-segment finally leaves the enzyme complex. Hydrolysis of the second ATP returns the system to the initial step of a cycle.
As the result of a catalytic cycle two ATP molecules are hydrolyzed and two negative supercoils are introduced into the DNA template. The number of superhelical turns introduced into an initially relaxed circular DNA has been calculated to be approximately equal to the number of ATP molecules hydrolyzed by gyrase Therefore, it can be suggested that two ATP molecules are hydrolyzed per cycle of reaction by gyrase, leading to the introduction of a linking difference of -2.
Gyrase specificity
Gyrase has a pronounced specificity to DNA substrates. Strong gyrase binding sites (SGS) were found in some phages (bacteriophage Mu
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus (the first of its kind to be identified) of the family ''Myoviridae'' which has been shown to cause genetic transposition. It is of particular importance as its discover ...
group) and plasmids (pSC101
pSC101 is a DNA plasmid that is used as a cloning vector in genetic cloning experiments. pSC101 was the first cloning vector, used in 1973 by Herbert Boyer and Stanley Norman Cohen. Using this plasmid they have demonstrated that a gene from a fro ...
, pBR322
pBR322 is a plasmid and was one of the first widely used ''E. coli'' cloning vectors. Created in 1977 in the laboratory of Herbert Boyer at the University of California, San Francisco, it was named after Francisco Bolivar Zapata, the postdoctoral ...
). Recently, high throughput mapping of DNA gyrase sites in the ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' genome using Topo-Seq approach revealed a long (≈130 bp) and degenerate binding motif that can explain the existence of SGSs. The gyrase motif reflects wrapping of DNA around the enzyme complex and DNA flexibility. It contains two periodic regions in which GC-rich islands are alternated with AT-rich patches by a period close to the period of DNA double helix (≈10.5 bp). The two regions correspond to DNA binding by C-terminal domains of GyrA subunits and resemble eukaryotic nucleosome binding motif.
Inhibition by antibiotics
Gyrase is present in prokaryotes and some eukaryotes, but the enzymes are not entirely similar in structure or sequence, and have different affinities for different molecules. This makes gyrase a good target forantibiotics
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention ...
. Two classes of antibiotics that inhibit gyrase are:
*The aminocoumarin
Aminocoumarin is a class of antibiotics that act by an inhibition of the DNA gyrase enzyme involved in the cell division in bacteria. They are derived from ''Streptomyces '' species, whose best-known representative – ''Streptomyces coelicolor ...
s (including novobiocin
Novobiocin, also known as albamycin or cathomycin, is an aminocoumarin antibiotic that is produced by the actinomycete '' Streptomyces niveus'', which has recently been identified as a subjective synonym for ''S. spheroides'' a member of the clas ...
and Coumermycin A1), which work by competitive inhibition
Competitive inhibition is interruption of a chemical pathway owing to one chemical substance inhibiting the effect of another by competing with it for binding or bonding. Any metabolic or chemical messenger system can potentially be affected ...
of energy transduction of DNA gyrase by binding to the ATPase active site on the GyrB subunit.
*The quinolones
Quinolone may refer to:
* 2-Quinolone
* 4-Quinolone
* Quinolone antibiotic
A quinolone antibiotic is a member of a large group of broad-spectrum bacteriocidals that share a bicyclic core structure related to the substance 4-quinolone. They a ...
(including nalidixic acid
Nalidixic acid (tradenames Nevigramon, NegGram, Wintomylon and WIN 18,320) is the first of the synthetic quinolone antibiotics.
In a technical sense, it is a naphthyridone, not a quinolone: its ring structure is a 1,8-naphthyridine nucleus that ...
and ciprofloxacin
Ciprofloxacin is a fluoroquinolone antibiotic used to treat a number of bacterial infections. This includes bone and joint infections, intra abdominal infections, certain types of infectious diarrhea, respiratory tract infections, skin infe ...
) are known as topoisomerase poisons. By binding to the enzyme they trap it in a transient step of the catalytic cycle, preventing the reunion of a G-segment. This results in an accumulation of double-strand break
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dama ...
s, stalling of replication forks and cell death. Quinolone-resistant bacteria frequently harbor mutated topoisomerases that resist quinolone binding.
The subunit A is selectively inactivated by antibiotics such as oxolinic and nalidixic acids. The subunit B is selectively inactivated by antibiotics such as coumermycin A1 and novobiocin. Inhibition of either subunit blocks supertwisting activity.
Phage T4
Phage T4gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
s 39, 52 and 60 encode proteins that form a DNA gyrase that is employed in phage DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance ...
during infection of the ''E. coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Esche ...
'' bacterial host.McCarthy D. Gyrase-dependent initiation of bacteriophage T4 DNA replication: interactions of Escherichia coli gyrase with novobiocin, coumermycin and phage DNA-delay gene products. J Mol Biol. 1979;127(3):265-283. doi:10.1016/0022-2836(79)90329-2 The phage gene 52 protein shares homology with the bacterial gyrase gyrA subunit and the phage gene 39 protein shares homology with the gyrB subunit. Since the host ''E. coli'' DNA gyrase can partially compensate for the loss of the phage gene products, mutants defective in either genes 39, 52 or 60 do not completely abolish phage DNA replication, but rather delay its initiation. Mutants defective in genes 39, 52 or 60 show increased genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryot ...
as well as increased base-substitution and deletion mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitos ...
suggesting that the host compensated DNA synthesis is less accurate than that directed by wild-type phage. A mutant defective in gene 39 also shows increased sensitivity to inactivation by ultraviolet
Ultraviolet (UV) is a form of electromagnetic radiation with wavelength from 10 nm (with a corresponding frequency around 30 PHz) to 400 nm (750 THz), shorter than that of visible light, but longer than X-rays. UV radiatio ...
irradiation during the stage of phage infection after initiation of DNA replication when multiple copies of the phage chromosome are present.Hyman P. The genetics of the Luria-Latarjet effect in bacteriophage T4: evidence for the involvement of multiple DNA repair pathways. Genet Res. 1993;62(1):1-9. doi:10.1017/s0016672300031499
See also
* GyrA RNA motifReferences
External links
* PDBe-KB : an overview of all the structure information available in the PDB for Escherichia coli DNA gyrase subunit A * PDBe-KB : an overview of all the structure information available in the PDB for Salmonella enterica DNA gyrase subunit B {{Portal bar, Biology, border=no DNA EC 5.99.1