alternative splicing on:
[Wikipedia]
[Google]
[Amazon]
 Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during

 Five basic modes of alternative splicing are generally recognized.
* Exon skipping or cassette exon: in this case, an
Five basic modes of alternative splicing are generally recognized.
* Exon skipping or cassette exon: in this case, an 
These modes describe basic splicing mechanisms, but may be inadequate to describe complex splicing events. For instance, the figure to the right shows 3 spliceforms from the mouse
 When the pre-mRNA has been transcribed from the DNA, it includes several
When the pre-mRNA has been transcribed from the DNA, it includes several
 Splicing is regulated by
Splicing is regulated by  In general, the determinants of splicing work in an inter-dependent manner that depends on context, so that the rules governing how splicing is regulated form a splicing code. The presence of a particular
In general, the determinants of splicing work in an inter-dependent manner that depends on context, so that the rules governing how splicing is regulated form a splicing code. The presence of a particular
 Pre-mRNAs from the ''D. melanogaster'' gene '' dsx'' contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a
Pre-mRNAs from the ''D. melanogaster'' gene '' dsx'' contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a
 Pre-mRNAs of the ''
Pre-mRNAs of the ''
 Multiple isoforms of the
Multiple isoforms of the

A General Definition and Nomenclature for Alternative Splicing Events
at
AStalavista (Alternative Splicing landscape visualization tool), a method for the computationally exhaustive classification of Alternative Splicing Structures
IsoPred: computationally predicted isoform functions
Stamms-lab.net: Research Group dealing with alternative Splicing issues and mis-splicing in human diseases
Alternative Splicing of ion channels in the brain, connected to mental and neurological diseases
BIPASS: Web Services in Alternative Splicing
{{DEFAULTSORT:Alternative Splicing Gene expression Spliceosome RNA splicing fr:Épissage#Épissage alternatif it:Splicing#Splicing alternativo
 Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. ...
that allows a single gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
to code for multiple protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
s. In this process, particular exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequenc ...
s of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequenc ...
s are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure).
Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bact ...
s, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants are due to splicing errors and the actual number of biologically relevant alternatively spliced genes is much lower.
There are numerous modes of alternative splicing observed, of which the most common is exon skipping
In molecular biology, exon skipping is a form of RNA splicing used to cause cells to “skip” over faulty or misaligned sections (exons) of genetic code, leading to a truncated but still functional protein despite the genetic mutation.
Mechanis ...
. In this mode, a particular exon may be included in mRNAs under some conditions or in particular tissues, and omitted from the mRNA in others.
The production of alternatively spliced mRNAs is regulated by a system of trans-acting In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regula ...
proteins that bind to cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
sites on the primary transcript
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs ...
itself. Such proteins include splicing activators that promote the usage of a particular splice site, and splicing repressors that reduce the usage of a particular site. Mechanisms of alternative splicing are highly variable, and new examples are constantly being found, particularly through the use of high-throughput techniques. Researchers hope to fully elucidate the regulatory systems involved in splicing, so that alternative splicing products from a given gene under particular conditions ("splicing variants") could be predicted by a "splicing code".
Abnormal variations in splicing are also implicated in disease; a large proportion of human genetic disorder
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosomal abnormality. Although polygenic disorde ...
s result from splicing variants. Abnormal splicing variants are also thought to contribute to the development of cancer, and splicing factor genes are frequently mutated in different types of cancer.
Discovery
Alternative splicing was first observed in 1977. TheAdenovirus
Adenoviruses (members of the family ''Adenoviridae'') are medium-sized (90–100 nm), nonenveloped (without an outer lipid bilayer) viruses with an icosahedral nucleocapsid containing a double-stranded DNA genome. Their name derives from t ...
produces five primary transcripts early in its infectious cycle, prior to viral DNA replication, and an additional one later, after DNA replication begins. The early primary transcripts continue to be produced after DNA replication begins. The additional primary transcript produced late in infection is large and comes from 5/6 of the 32kb adenovirus genome. This is much larger than any of the individual adenovirus
Adenoviruses (members of the family ''Adenoviridae'') are medium-sized (90–100 nm), nonenveloped (without an outer lipid bilayer) viruses with an icosahedral nucleocapsid containing a double-stranded DNA genome. Their name derives from t ...
mRNAs present in infected cells. Researchers found that the primary RNA transcript produced by adenovirus type 2 in the late phase was spliced in many different ways, resulting in mRNAs encoding different viral proteins. In addition, the primary transcript contained multiple polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eu ...
sites, giving different 3’ ends for the processed mRNAs.
In 1981, the first example of alternative splicing in a transcript from a normal, endogenous
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell.
In contrast, exogenous substances and processes are those that originate from outside of an organism.
For example, ...
gene was characterized. The gene encoding the thyroid
The thyroid, or thyroid gland, is an endocrine gland in vertebrates. In humans it is in the neck and consists of two connected lobes. The lower two thirds of the lobes are connected by a thin band of tissue called the thyroid isthmus. The t ...
hormone calcitonin
Calcitonin is a 32 amino acid peptide hormone secreted by parafollicular cells (also known as C cells) of the thyroid (or endostyle) in humans and other chordates. in the ultimopharyngeal body. It acts to reduce blood calcium (Ca2+), opposing ...
was found to be alternatively spliced in mammalian cells. The primary transcript from this gene contains 6 exons; the calcitonin
Calcitonin is a 32 amino acid peptide hormone secreted by parafollicular cells (also known as C cells) of the thyroid (or endostyle) in humans and other chordates. in the ultimopharyngeal body. It acts to reduce blood calcium (Ca2+), opposing ...
mRNA contains exons 1–4, and terminates after a polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eu ...
site in exon 4. Another mRNA is produced from this pre-mRNA by skipping exon 4, and includes exons 1–3, 5, and 6. It encodes a protein known as CGRP (calcitonin gene related peptide
Calcitonin gene-related peptide (CGRP) is a member of the calcitonin family of peptides consisting of calcitonin, amylin, adrenomedullin, adrenomedullin 2 (intermedin) and calcitonin‑receptor‑stimulating peptide. Calcitonin is mainly produce ...
). Examples of alternative splicing in immunoglobin gene transcripts in mammals were also observed in the early 1980s.
Since then, many other examples of biologically relevant alternative splicing have been found in eukaryotes. The "record-holder" for alternative splicing is a ''D. melanogaster'' gene called Dscam, which could potentially have 38,016 splice variants.
In 2021, it was discovered that the genome of adenovirus type 2, the adenovirus in which alternative splicing was first identified, was able to produce a much greater variety of mRNA than previously thought. By using next generation sequencing technology, researchers were able to update the human adenovirus type 2 transcriptome, and present a mind-boggling 904 unique mRNA, produced by the virus through a complex pattern of alternative splicing. Very few of these splice variants have been shown to be functional, a point that the authors raise in their paper.
::"An outstanding question is what roles the menagerie of novel RNAs play or whether they are spurious molecules generated by an overloaded splicing machinery."
Modes

 Five basic modes of alternative splicing are generally recognized.
* Exon skipping or cassette exon: in this case, an
Five basic modes of alternative splicing are generally recognized.
* Exon skipping or cassette exon: in this case, an exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequenc ...
may be spliced out of the primary transcript or retained. This is the most common mode in mammalian pre-mRNA
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs ...
s.
* Mutually exclusive exons: One of two exons is retained in mRNAs after splicing, but not both.
* Alternative donor site: An alternative 5' splice junction (donor site) is used, changing the 3' boundary of the upstream
Upstream may refer to:
* Upstream (bioprocess)
* ''Upstream'' (film), a 1927 film by John Ford
* Upstream (networking)
* ''Upstream'' (newspaper), a newspaper covering the oil and gas industry
* Upstream (petroleum industry)
* Upstream (software ...
exon.
* Alternative acceptor site: An alternative 3' splice junction (acceptor site) is used, changing the 5' boundary of the downstream exon.
* Intron retention: A sequence may be spliced out as an intron or simply retained. This is distinguished from exon skipping because the retained sequence is not flanked by introns
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene ...
. If the retained intron is in the coding region, the intron must encode amino acids in frame with the neighboring exons, or a stop codon or a shift in the reading frame
In molecular biology, a reading frame is a way of dividing the sequence of nucleotides in a nucleic acid ( DNA or RNA) molecule into a set of consecutive, non-overlapping triplets. Where these triplets equate to amino acids or stop signals during ...
will cause the protein to be non-functional. This is the rarest mode in mammals but the most common in plants.
In addition to these primary modes of alternative splicing, there are two other main mechanisms by which different mRNAs may be generated from the same gene; multiple promoters and multiple polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eu ...
sites. Use of multiple promoters is properly described as a transcriptional regulation
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA ( transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from al ...
mechanism rather than alternative splicing; by starting transcription at different points, transcripts with different 5'-most exons can be generated. At the other end, multiple polyadenylation sites provide different 3' end points for the transcript. Both of these mechanisms are found in combination with alternative splicing and provide additional variety in mRNAs derived from a gene. 
These modes describe basic splicing mechanisms, but may be inadequate to describe complex splicing events. For instance, the figure to the right shows 3 spliceforms from the mouse
hyaluronidase
Hyaluronidases are a family of enzymes that catalyse the degradation of hyaluronic acid (HA). Karl Meyer classified these enzymes in 1971, into three distinct groups, a scheme based on the enzyme reaction products. The three main types of hyalu ...
3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows intron retention, whereas the comparison between the second and the third spliceform (yellow vs. blue) exhibits exon skipping. A model nomenclature to uniquely designate all possible splicing patterns has recently been proposed.
Mechanisms
General splicing mechanism
 When the pre-mRNA has been transcribed from the DNA, it includes several
When the pre-mRNA has been transcribed from the DNA, it includes several intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene ...
s and exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequenc ...
s. (In nematodes, the mean is 4–5 exons and introns; in the fruit fly ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many s ...
'' there can be more than 100 introns and exons in one transcribed pre-mRNA.) The exons to be retained in the mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
are determined during the splicing process. The regulation and selection of splice sites are done by trans-acting splicing activator and splicing repressor proteins as well as cis-acting elements within the pre-mRNA itself such as exonic splicing enhancers and exonic splicing silencers.
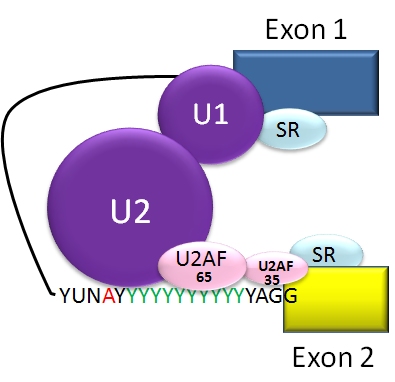
The typical eukaryotic nuclear intron has consensus sequences defining important regions. Each intron has the sequence GU at its 5' end. Near the 3' end there is a branch site. The nucleotide at the branchpoint is always an A; the consensus around this sequence varies somewhat. In humans the branch site consensus sequence is yUnAy. The branch site is followed by a series of pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The othe ...
s – the polypyrimidine tract {{Short description, A pyrimidine-rich sequence involved in pre-messenger RNA maturation
The polypyrimidine tract is a region of pre-messenger RNA (mRNA) that promotes the assembly of the spliceosome, the protein complex specialized for carrying ou ...
– then by AG at the 3' end.
Splicing of mRNA is performed by an RNA and protein complex known as the spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
, containing snRNP
snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pr ...
s designated U1, U2, U4, U5, and U6 (U3 is not involved in mRNA splicing). U1 binds to the 5' GU and U2, with the assistance of the U2AF
U or u, is the twenty-first and sixth-to-last letter and fifth vowel letter of the Latin alphabet, used in the modern English alphabet, the alphabets of other western European languages and others worldwide. Its name in English is ''u'' (pr ...
protein factors, binds to the branchpoint A within the branch site. The complex at this stage is known as the spliceosome A complex. Formation of the A complex is usually the key step in determining the ends of the intron to be spliced out, and defining the ends of the exon to be retained. (The U nomenclature derives from their high uridine content).
The U4,U5,U6 complex binds, and U6 replaces the U1 position. U1 and U4 leave. The remaining complex then performs two transesterification
In organic chemistry, transesterification is the process of exchanging the organic group R″ of an ester with the organic group R' of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst. The reaction ca ...
reactions. In the first transesterification, 5' end of the intron is cleaved from the upstream exon and joined to the branch site A by a 2',5'-phosphodiester
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is ...
linkage. In the second transesterification, the 3' end of the intron is cleaved from the downstream exon, and the two exons are joined by a phosphodiester bond. The intron is then released in lariat form and degraded.
Regulatory elements and proteins
 Splicing is regulated by
Splicing is regulated by trans-acting In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regula ...
proteins (repressors and activators) and corresponding cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
regulatory sites (silencers and enhancers) on the pre-mRNA. However, as part of the complexity of alternative splicing, it is noted that the effects of a splicing factor are frequently position-dependent. That is, a splicing factor that serves as a splicing activator when bound to an intronic enhancer element may serve as a repressor when bound to its splicing element in the context of an exon, and vice versa. The secondary structure of the pre-mRNA transcript also plays a role in regulating splicing, such as by bringing together splicing elements or by masking a sequence that would otherwise serve as a binding element for a splicing factor. Together, these elements form a "splicing code" that governs how splicing will occur under different cellular conditions.
There are two major types of cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
RNA sequence elements present in pre-mRNAs and they have corresponding trans-acting In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regula ...
RNA-binding protein
RNA-binding proteins (often abbreviated as RBPs) are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes.
RBPs contain various structural motifs, such as RNA recognition moti ...
s. Splicing ''silencers'' are sites to which splicing repressor proteins bind, reducing the probability that a nearby site will be used as a splice junction. These can be located in the intron itself (intronic splicing silencers, ISS) or in a neighboring exon ( exonic splicing silencers, ESS). They vary in sequence, as well as in the types of proteins that bind to them. The majority of splicing repressors are heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNPA1 and polypyrimidine tract binding protein (PTB). Splicing ''enhancers'' are sites to which splicing activator proteins bind, increasing the probability that a nearby site will be used as a splice junction. These also may occur in the intron (intronic splicing enhancers, ISE) or exon ( exonic splicing enhancers, ESE). Most of the activator proteins that bind to ISEs and ESEs are members of the SR protein family. Such proteins contain RNA recognition motifs and arginine and serine-rich (RS) domains.
 In general, the determinants of splicing work in an inter-dependent manner that depends on context, so that the rules governing how splicing is regulated form a splicing code. The presence of a particular
In general, the determinants of splicing work in an inter-dependent manner that depends on context, so that the rules governing how splicing is regulated form a splicing code. The presence of a particular cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
RNA sequence element may increase the probability that a nearby site will be spliced in some cases, but decrease the probability in other cases, depending on context. The context within which regulatory elements act includes cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
context that is established by the presence of other RNA sequence features, and trans-acting In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regula ...
context that is established by cellular conditions. For example, some cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
RNA sequence elements influence splicing only if multiple elements are present in the same region so as to establish context. As another example, a cis-acting
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphog ...
element can have opposite effects on splicing, depending on which proteins are expressed in the cell (e.g., neuronal versus non-neuronal PTB). The adaptive significance of splicing silencers and enhancers is attested by studies showing that there is strong selection in human genes against mutations that produce new silencers or disrupt existing enhancers.
DNA methylation and alternative splicing in social insects
CpG DNA methylation has showed a role to regulate the alternative splicing in social insects. In honey bees (''Apis mellifera''), CpG DNA methylation seems to regulate the exon skipping based on the first few genomic studies after honey bee genome was available. CpG DNA methylation regulated alternative splicing more extensively, not only affect exon skipping, but also intron retention, and other splicing events.Examples
Exon skipping: ''Drosophila'' ''dsx''
 Pre-mRNAs from the ''D. melanogaster'' gene '' dsx'' contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a
Pre-mRNAs from the ''D. melanogaster'' gene '' dsx'' contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eu ...
signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development.
This is an example of exon skipping. The intron upstream from exon 4 has a polypyrimidine tract {{Short description, A pyrimidine-rich sequence involved in pre-messenger RNA maturation
The polypyrimidine tract is a region of pre-messenger RNA (mRNA) that promotes the assembly of the spliceosome, the protein complex specialized for carrying ou ...
that doesn't match the consensus sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified r ...
well, so that U2AF proteins bind poorly to it without assistance from splicing activators. This 3' splice acceptor site is therefore not used in males. Females, however, produce the splicing activator Transformer (Tra) (see below). The SR protein Tra2 is produced in both sexes and binds to an ESE in exon 4; if Tra is present, it binds to Tra2 and, along with another SR protein, forms a complex that assists U2AF proteins in binding to the weak polypyrimidine tract. U2 is recruited to the associated branchpoint, and this leads to inclusion of exon 4 in the mRNA.
Alternative acceptor sites: ''Drosophila'' '
 Pre-mRNAs of the ''
Pre-mRNAs of the ''Transformer
A transformer is a passive component that transfers electrical energy from one electrical circuit to another circuit, or multiple circuits. A varying current in any coil of the transformer produces a varying magnetic flux in the transformer' ...
'' (Tra) gene of ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the " vinegar fly" or " pomace fly". Starting with ...
'' undergo alternative splicing via the alternative acceptor site mode. The gene Tra encodes a protein that is expressed only in females. The primary transcript of this gene contains an intron with two possible acceptor sites. In males, the upstream acceptor site is used. This causes a longer version of exon 2 to be included in the processed transcript, including an early stop codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon ( nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in ...
. The resulting mRNA encodes a truncated protein product that is inactive. Females produce the master sex determination protein Sex lethal (Sxl). The Sxl protein is a splicing repressor that binds to an ISS in the RNA of the Tra transcript near the upstream acceptor site, preventing U2AF
U or u, is the twenty-first and sixth-to-last letter and fifth vowel letter of the Latin alphabet, used in the modern English alphabet, the alphabets of other western European languages and others worldwide. Its name in English is ''u'' (pr ...
protein from binding to the polypyrimidine tract. This prevents the use of this junction, shifting the spliceosome binding to the downstream acceptor site. Splicing at this point bypasses the stop codon, which is excised as part of the intron. The resulting mRNA encodes an active Tra protein, which itself is a regulator of alternative splicing of other sex-related genes (see ''dsx'' above).
Exon definition: Fas receptor
 Multiple isoforms of the
Multiple isoforms of the Fas receptor
The Fas receptor, also known as Fas, FasR, apoptosis antigen 1 (APO-1 or APT), cluster of differentiation 95 (CD95) or tumor necrosis factor receptor superfamily member 6 (TNFRSF6), is a protein that in humans is encoded by the ''FAS'' gene. Fas ...
protein are produced by alternative splicing. Two normally occurring isoforms in humans are produced by an exon-skipping mechanism. An mRNA including exon 6 encodes the membrane-bound form of the Fas receptor, which promotes apoptosis, or programmed cell death. Increased expression of Fas receptor in skin cells chronically exposed to the sun, and absence of expression in skin cancer cells, suggests that this mechanism may be important in elimination of pre-cancerous cells in humans. If exon 6 is skipped, the resulting mRNA encodes a soluble Fas protein that does not promote apoptosis. The inclusion or skipping of the exon depends on two antagonistic proteins, TIA-1
TIA1 or Tia1 cytotoxic granule-associated rna binding protein is a 3'UTR mRNA binding protein that can bind the 5'TOP sequence of 5'TOP mRNAs. It is associated with programmed cell death ( apoptosis) and regulates alternative splicing of the ge ...
and polypyrimidine tract-binding protein (PTB).
* The 5' donor site in the intron downstream from exon 6 in the pre-mRNA has a weak agreement with the consensus sequence, and is not bound usually by the U1 snRNP. If U1 does not bind, the exon is skipped (see "a" in accompanying figure).
* Binding of TIA-1 protein to an intronic splicing enhancer site stabilizes binding of the U1 snRNP. The resulting 5' donor site complex assists in binding of the splicing factor U2AF to the 3' splice site upstream of the exon, through a mechanism that is not yet known (see b).
* Exon 6 contains a pyrimidine-rich exonic splicing silencer, ''ure6'', where PTB can bind. If PTB binds, it inhibits the effect of the 5' donor complex on the binding of U2AF to the acceptor site, resulting in exon skipping (see c).
This mechanism is an example of exon definition in splicing. A spliceosome assembles on an intron, and the snRNP subunits fold the RNA so that the 5' and 3' ends of the intron are joined. However, recently studied examples such as this one show that there are also interactions between the ends of the exon. In this particular case, these exon definition interactions are necessary to allow the binding of core splicing factors prior to assembly of the spliceosomes on the two flanking introns.
Repressor-activator competition: HIV-1 ''tat'' exon 2

HIV
The human immunodeficiency viruses (HIV) are two species of '' Lentivirus'' (a subgroup of retrovirus) that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immu ...
, the retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase ...
that causes AIDS in humans, produces a single primary RNA transcript, which is alternatively spliced in multiple ways to produce over 40 different mRNAs. Equilibrium among differentially spliced transcripts provides multiple mRNAs encoding different products that are required for viral multiplication. One of the differentially spliced transcripts contains the ''tat'' gene, in which exon 2 is a cassette exon that may be skipped or included. The inclusion of tat exon 2 in the RNA is regulated by competition between the splicing repressor hnRNP A1 and the SR protein SC35. Within exon 2 an exonic splicing silencer sequence (ESS) and an exonic splicing enhancer sequence (ESE) overlap. If A1 repressor protein binds to the ESS, it initiates cooperative binding of multiple A1 molecules, extending into the 5’ donor site upstream of exon 2 and preventing the binding of the core splicing factor U2AF35 to the polypyrimidine tract. If SC35 binds to the ESE, it prevents A1 binding and maintains the 5’ donor site in an accessible state for assembly of the spliceosome. Competition between the activator and repressor ensures that both mRNA types (with and without exon 2) are produced.
Adaptive significance
Genuine alternative splicing occurs in both protein-coding genes and non-coding genes to produce multiple products (proteins or non-coding RNAs). External information is needed in order to decide which product is made, given a DNA sequence and the initial transcript. Since the methods of regulation are inherited, this provides novel ways for mutations to affect gene expression. Alternative splicing may provide evolutionary flexibility. A single point mutation may cause a given exon to be occasionally excluded or included from a transcript during splicing, allowing production of a newprotein isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some iso ...
without loss of the original protein. Studies have identified intrinsically disordered regions (see Intrinsically unstructured proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs rang ...
) as enriched in the non-constitutive exons suggesting that protein isoforms may display functional diversity due to the alteration of functional modules within these regions. Such functional diversity achieved by isoforms is reflected by their expression patterns and can be predicted by machine learning approaches. Comparative studies indicate that alternative splicing preceded multicellularity in evolution, and suggest that this mechanism might have been co-opted to assist in the development of multicellular organisms.
Research based on the Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
and other genome sequencing has shown that humans have only about 30% more genes than the roundworm ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (r ...
'', and only about twice as many as the fly ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the " vinegar fly" or " pomace fly". Starting with ...
''. This finding led to speculation that the perceived greater complexity of humans, or vertebrates generally, might be due to higher rates of alternative splicing in humans than are found in invertebrates.
However, a study on samples of 100,000 ESTs each from human, mouse, rat, cow, fly (''D. melanogaster''), worm (''C. elegans''), and the plant ''Arabidopsis thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small flowering plant native to Eurasia and Africa. ''A. thaliana'' is considered a weed; it is found along the shoulders of roads and in disturbed land.
A winter ...
'' found no large differences in frequency of alternatively spliced genes among humans and any of the other animals tested. Another study, however, proposed that these results were an artifact of the different numbers of ESTs available for the various organisms. When they compared alternative splicing frequencies in random subsets of genes from each organism, the authors concluded that vertebrates do have higher rates of alternative splicing than invertebrates.
Disease
Changes in the RNA processing machinery may lead to mis-splicing of multiple transcripts, while single-nucleotide alterations in splice sites or cis-acting splicing regulatory sites may lead to differences in splicing of a single gene, and thus in the mRNA produced from a mutant gene's transcripts. A study in 2005 involving probabilistic analyses indicated that greater than 60% of human disease-causingmutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
s affect splicing rather than directly affecting coding sequences. A more recent study indicates that one-third of all hereditary diseases are likely to have a splicing component. Regardless of exact percentage, a number of splicing-related diseases do exist. As described below, a prominent example of splicing-related diseases is cancer.
Abnormally spliced mRNAs are also found in a high proportion of cancerous cells. Combined RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing ...
and proteomics analyses have revealed striking differential expression
of splice isoforms of key proteins in important cancer pathways. It is not always clear whether such aberrant patterns of splicing contribute to the cancerous growth, or are merely consequence of cellular abnormalities associated with cancer. For certain types of cancer, like in colorectal and prostate, the number of splicing errors per cancer has been shown to vary greatly between individual cancers, a phenomenon referred to as transcriptome instability. Transcriptome instability has further been shown to correlate grealty with reduced expression level of splicing factor genes. Mutation of ''DNMT3A
DNA (cytosine-5)-methyltransferase 3A is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.
This enzyme is responsible f ...
'' has been demonstrated to contribute to hematologic malignancies
Tumors of the hematopoietic and lymphoid tissues (American English) or tumours of the haematopoietic and lymphoid tissues (British English) are tumors that affect the blood, bone marrow, lymph, and lymphatic system. Because these tissues are all ...
, and that ''DNMT3A
DNA (cytosine-5)-methyltransferase 3A is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.
This enzyme is responsible f ...
''-mutated cell lines exhibit transcriptome instability as compared to their isogenic wildtype counterparts.
In fact, there is actually a reduction of alternative splicing in cancerous cells compared to normal ones, and the types of splicing differ; for instance, cancerous cells show higher levels of intron retention than normal cells, but lower levels of exon skipping. Some of the differences in splicing in cancerous cells may be due to the high frequency of somatic mutations in splicing factor genes, and some may result from changes in phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, ...
of trans-acting splicing factors. Others may be produced by changes in the relative amounts of splicing factors produced; for instance, breast cancer cells have been shown to have increased levels of the splicing factor SF2/ASF. One study found that a relatively small percentage (383 out of over 26000) of alternative splicing variants were significantly higher in frequency in tumor cells than normal cells, suggesting that there is a limited set of genes which, when mis-spliced, contribute to tumor development. It is believed however that the deleterious effects of mis-spliced transcripts are usually safeguarded and eliminated by a cellular posttranscriptional quality control mechanism termed Nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant ...
MD
One example of a specific splicing variant associated with cancers is in one of the human DNMT genes. Three DNMT genes encode enzymes that add methyl groups to DNA, a modification that often has regulatory effects. Several abnormally spliced DNMT3B mRNAs are found in tumors and cancer cell lines. In two separate studies, expression of two of these abnormally spliced mRNAs in mammalian cells caused changes in the DNA methylation patterns in those cells. Cells with one of the abnormal mRNAs also grew twice as fast as control cells, indicating a direct contribution to tumor development by this product.
Another example is the ''Ron'' ('' MST1R'') proto-oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.
. An important property of cancerous cells is their ability to move and invade normal tissue. Production of an abnormally spliced transcript of ''Ron'' has been found to be associated with increased levels of the SF2/ASF in breast cancer cells. The abnormal isoform of the Ron protein encoded by this mRNA leads to cell motility
Motility is the ability of an organism to move independently, using metabolic energy.
Definitions
Motility, the ability of an organism to move independently, using metabolic energy, can be contrasted with sessility, the state of organisms th ...
.
Overexpression of a truncated splice variant of the FOSB
Protein fosB, also known as FosB and G0/G1 switch regulatory protein 3 (G0S3), is a protein that in humans is encoded by the FBJ murine osteosarcoma viral oncogene homolog B (''FOSB'') gene.
The FOS gene family consists of four members: FOS, F ...
gene – ΔFosB
Protein fosB, also known as FosB and G0/G1 switch regulatory protein 3 (G0S3), is a protein that in humans is encoded by the FBJ murine osteosarcoma viral oncogene homolog B (''FOSB'') gene.
The FOS gene family consists of four members: FOS, F ...
– in a specific population of neurons in the nucleus accumbens
The nucleus accumbens (NAc or NAcc; also known as the accumbens nucleus, or formerly as the ''nucleus accumbens septi'', Latin for "nucleus adjacent to the septum") is a region in the basal forebrain rostral to the preoptic area of the hypot ...
has been identified as the causal mechanism involved in the induction and maintenance of an addiction
Addiction is a neuropsychological disorder characterized by a persistent and intense urge to engage in certain behaviors, one of which is the usage of a drug, despite substantial harm and other negative consequences. Repetitive drug use oft ...
to drugs and natural rewards.
Recent provocative studies point to a key function of chromatin structure and histone modifications in alternative splicing regulation. These insights suggest that epigenetic regulation determines not only what parts of the genome are expressed but also how they are spliced.
Genome-wide analysis
Genome-wide analysis of alternative splicing is a challenging task. Typically, alternatively spliced transcripts have been found by comparing EST sequences, but this requires sequencing of very large numbers of ESTs. Most EST libraries come from a very limited number of tissues, so tissue-specific splice variants are likely to be missed in any case. High-throughput approaches to investigate splicing have, however, been developed, such as:DNA microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to ...
-based analyses, RNA-binding assays, and deep sequencing
Coverage (or depth) in DNA sequencing is the number of unique reads that include a given nucleotide in the reconstructed sequence. Deep sequencing refers to the general concept of aiming for high number of unique reads of each region of a sequence ...
. These methods can be used to screen for polymorphisms or mutations in or around splicing elements that affect protein binding. When combined with splicing assays, including ''in vivo'' reporter gene
In molecular biology, a reporter gene (often simply reporter) is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the char ...
assays, the functional effects of polymorphisms or mutations on the splicing of pre-mRNA transcripts can then be analyzed.
In microarray analysis, arrays of DNA fragments representing individual exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequenc ...
s (''e.g.'' Affymetrix
Affymetrix is now Applied Biosystems, a brand of DNA microarray products sold by Thermo Fisher Scientific that originated with an American biotechnology research and development and manufacturing company of the same name. The Santa Clara, Califor ...
exon microarray) or exon/exon boundaries (''e.g.'' arrays from ExonHit or Jivan Jivan may refer to:
* Jivan Avetisyan, Armenian film director
* Bhai Jivan Singh
* Alin Jivan, Romanian artist gymnast
* Kantilal Jivan
* Bhanwarlal Jivan, Indian politician
* Jag Jivan Ram, Indian politician
* Jivan Gasparyan, Armenian musicia ...
) have been used. The array is then probed with labeled cDNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a speci ...
from tissues of interest. The probe cDNAs bind to DNA from the exons that are included in mRNAs in their tissue of origin, or to DNA from the boundary where two exons have been joined. This can reveal the presence of particular alternatively spliced mRNAs.
CLIP (Cross-link
In chemistry and biology a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural ...
ing and immunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sam ...
) uses UV radiation to link proteins to RNA molecules in a tissue during splicing. A trans-acting splicing regulatory protein of interest is then precipitated using specific antibodies. When the RNA attached to that protein is isolated and cloned, it reveals the target sequences for that protein. Another method for identifying RNA-binding proteins and mapping their binding to pre-mRNA transcripts is "Microarray Evaluation of Genomic Aptamers by shift (MEGAshift)".net This method involves an adaptation of the "Systematic Evolution of Ligands by Exponential Enrichment (SELEX)" method together with a microarray-based readout. Use of the MEGAshift method has provided insights into the regulation of alternative splicing by allowing for the identification of sequences in pre-mRNA transcripts surrounding alternatively spliced exons that mediate binding to different splicing factors, such as ASF/SF2 and PTB. This approach has also been used to aid in determining the relationship between RNA secondary structure and the binding of splicing factors.
Deep sequencing technologies have been used to conduct genome-wide analyses of mRNAs – unprocessed and processed – thus providing insights into alternative splicing. For example, results from use of deep sequencing indicate that, in humans, an estimated 95% of transcripts from multiexon genes undergo alternative splicing, with a number of pre-mRNA transcripts spliced in a tissue-specific manner. Functional genomics and computational approaches based on multiple instance learning have also been developed to integrate RNA-seq data to predict functions for alternatively spliced isoforms. Deep sequencing has also aided in the ''in vivo'' detection of the transient lariats
A lasso ( or ), also called lariat, riata, or reata (all from Castilian, la reata 're-tied rope'), is a loop of rope designed as a restraint to be thrown around a target and tightened when pulled. It is a well-known tool of the Spanish an ...
that are released during splicing, the determination of branch site sequences, and the large-scale mapping of branchpoints in human pre-mRNA transcripts.
Use of reporter assays makes it possible to find the splicing proteins involved in a specific alternative splicing event by constructing reporter genes that will express one of two different fluorescent proteins depending on the splicing reaction that occurs. This method has been used to isolate mutants affecting splicing and thus to identify novel splicing regulatory proteins inactivated in those mutants.
Databases
There is a collection of alternative splicing databases. These databases are useful for finding genes having pre-mRNAs undergoing alternative splicing and alternative splicing events or to study the functional impact of alternative splicing.See also
* AspicDB database * Exitron * Intronerator * Polyadenylation § Alternative polyadenylation * ProSAS database * SpliceInfo database *Trans-splicing
''Trans''-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archae ...
References
External links
A General Definition and Nomenclature for Alternative Splicing Events
at
SciVee
SciVee http://www.scivee.tv scivee.tv was a science video sharing website where researchers could upload, view and share science video clips and connect them to scientific literature, posters and slides from 2007 to 2015. The SciVee website is ...
AStalavista (Alternative Splicing landscape visualization tool), a method for the computationally exhaustive classification of Alternative Splicing Structures
IsoPred: computationally predicted isoform functions
Stamms-lab.net: Research Group dealing with alternative Splicing issues and mis-splicing in human diseases
Alternative Splicing of ion channels in the brain, connected to mental and neurological diseases
BIPASS: Web Services in Alternative Splicing
{{DEFAULTSORT:Alternative Splicing Gene expression Spliceosome RNA splicing fr:Épissage#Épissage alternatif it:Splicing#Splicing alternativo