Prokaryotic DNA Replication on:
[Wikipedia]
[Google]
[Amazon]
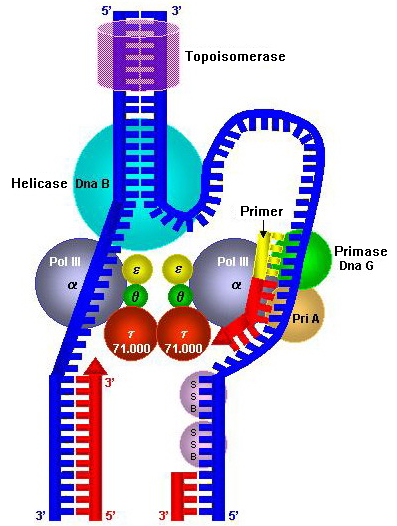 Once priming is complete,
Once priming is complete,
 . When conjugation is initiated by a signal the relaxase
. When conjugation is initiated by a signal the relaxase
Prokaryotic
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ...
DNA Replication is the process by which a prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism '' E. coli'', other bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
show many similarities. Replication is bi-directional and originates at a single origin of replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by se ...
(OriC). It consists of three steps: Initiation, elongation, and termination.

Initiation
All cells must finish DNA replication before they can proceed for cell division. Media conditions that support fast growth in bacteria also couples with shorter inter-initiation time in them, i.e. the doubling time in fast growing cells is less as compared to the slow growth. In other words, it is possible that in fast growth conditions the grandmother cells starts replicating its DNA for grand daughter cell. For the same reason, the initiation of DNA replication is highly regulated. Bacterial origins regulate orisome assembly, a nuclei-protein complex assembled on the origin responsible for unwinding the origin and loading all the replication machinery. In ''E. coli,'' the direction for orisome assembly are built into a short stretch of nucleotide sequence called as origin of replication (''oriC'') which contains multiple binding sites for the initiator protein DnaA (a highly homologous protein amongst bacterial kingdom). DnaA has four domains with each domain responsible for a specific task. There are 11 DnaA binding sites/boxes on the ''E. coli'' origin of replication out of which three boxes R1, R2 and R4 (which have a highly conserved 9 bp consensus sequence 5' - TTATC/ACACA ) are high affinity DnaA boxes. They bind to DnaA-ADP and DnaA-ATP with equal affinities and are bound by DnaA throughout most of the cell cycle and forms a scaffold on which rest of the orisome assembles. The rest eight DnaA boxes are low affinity sites that preferentially bind to DnaA-ATP. During initiation, DnaA bound to high affinity DnaA box R4 donates additional DnaA to the adjacent low affinity site and progressively fill all the low affinity DnaA boxes. Filling of the sites changes origin conformation from its native state. It is hypothesized that DNA stretching by DnaA bound to the origin promotes strand separation which allows more DnaA to bind to the unwound region. The DnaC helicase loader then interacts with the DnaA bound to the single-stranded DNA to recruit theDnaB helicase
DnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the qu ...
, which will continue to unwind the DNA as the DnaG primase lays down an RNA primer and DNA Polymerase III holoenzyme
DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. The complex has high processivity (i.e. the number of ...
begins elongation.
Regulation
Chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
replication in bacteria is regulated at the initiation stage. DnaA-ATP is hydrolyzed
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysis ...
into the inactive DnaA-ADP by RIDA (Regulatory Inactivation of DnaA), and converted back to the active DnaA-ATP form by DARS (DnaA Reactivating Sequence, which is itself regulated by Fis and IHF). However, the main source of DnaA-ATP is synthesis of new molecules. Meanwhile, several other proteins interact directly with the ''oriC'' sequence to regulate initiation, usually by inhibition. In ''E. coli'' these proteins include DiaA, SeqA, IciA, HU, and ArcA-P, but they vary across other bacterial species. A few other mechanisms in ''E. coli'' that variously regulate initiation are DDAH (''datA''-Dependent DnaA Hydrolysis, which is also regulated by IHF), inhibition of the ''dnaA'' gene (by the SeqA protein), and reactivation of DnaA by the lipid membrane.
Elongation
 Once priming is complete,
Once priming is complete, DNA polymerase III holoenzyme
DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. The complex has high processivity (i.e. the number of ...
is loaded into the DNA and replication begins. The catalytic mechanism of DNA polymerase III involves the use of two metal ions in the active site, and a region in the active site that can discriminate between deoxyribonucleotide
A deoxyribonucleotide is a nucleotide that contains deoxyribose. They are the monomeric units of the informational biopolymer, deoxyribonucleic acid ( DNA). Each deoxyribonucleotide comprises three parts: a deoxyribose sugar ( monosaccharide), a ni ...
s and ribonucleotide
In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic m ...
s. The metal ions are general divalent cations that help the 3' OH initiate a nucleophilic attack onto the alpha phosphate
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid .
The phosphate or orthophosphate ion is derived from phosph ...
of the deoxyribonucleotide and orient and stabilize the negatively charged triphosphate on the deoxyribonucleotide. Nucleophilic attack by the 3' OH on the alpha phosphate releases pyrophosphate
In chemistry, pyrophosphates are phosphorus oxyanions that contain two phosphorus atoms in a P–O–P linkage. A number of pyrophosphate salts exist, such as disodium pyrophosphate (Na2H2P2O7) and tetrasodium pyrophosphate (Na4P2O7), among o ...
, which is then subsequently hydrolyzed (by inorganic phosphatase) into two phosphates. This hydrolysis drives DNA synthesis to completion.
Furthermore, DNA polymerase III must be able to distinguish between correctly paired bases and incorrectly paired bases. This is accomplished by distinguishing Watson-Crick base pairs through the use of an active site pocket that is complementary in shape to the structure of correctly paired nucleotides. This pocket has a tyrosine residue that is able to form van der Waals interactions
A van is a type of road vehicle used for transporting goods or people. Depending on the type of van, it can be bigger or smaller than a pickup truck and SUV, and bigger than a common car. There is some varying in the scope of the word across th ...
with the correctly paired nucleotide. In addition, dsDNA (double stranded DNA) in the active site has a wider major groove
Major (commandant in certain jurisdictions) is a military rank of commissioned officer status, with corresponding ranks existing in many military forces throughout the world. When used unhyphenated and in conjunction with no other indicators ...
and shallower minor groove that permits the formation of hydrogen bonds with the third nitrogen
Nitrogen is the chemical element with the symbol N and atomic number 7. Nitrogen is a nonmetal and the lightest member of group 15 of the periodic table, often called the pnictogens. It is a common element in the universe, estimated at seve ...
of purine
Purine is a heterocyclic aromatic organic compound that consists of two rings ( pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purin ...
bases and the second oxygen
Oxygen is the chemical element with the symbol O and atomic number 8. It is a member of the chalcogen group in the periodic table, a highly reactive nonmetal, and an oxidizing agent that readily forms oxides with most elements as we ...
of pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The othe ...
bases. Finally, the active site makes extensive hydrogen bonds with the DNA backbone. These interactions result in the DNA polymerase III closing around a correctly paired base. If a base is inserted and incorrectly paired, these interactions could not occur due to disruptions in hydrogen bonding and van der Waals interactions.
DNA is read in the 3' → 5' direction, therefore, nucleotides are synthesized (or attached to the template strand) in the 5' → 3' direction. However, one of the parent strands of DNA is 3' → 5' while the other is 5' → 3'. To solve this, replication occurs in opposite directions. Heading towards the replication fork, the leading strand
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
is synthesized in a continuous fashion, only requiring one primer. On the other hand, the lagging strand
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inherita ...
, heading away from the replication fork, is synthesized in a series of short fragments known as Okazaki fragments, consequently requiring many primers. The RNA primers of Okazaki fragments are subsequently degraded by RNase H and DNA Polymerase I
DNA polymerase I (or Pol I) is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase (and the first known of any kind of polymerase). It was initial ...
(exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is t ...
), and the gaps (or nicks
Nix (or Nicks) is a surname of English origin, which initially indicated that the person so named was the child of a person named Nicholas, traditionally shortened to "Nick". It is therefore closely related to Nixon and Nickson, which are derive ...
) are filled with deoxyribonucleotides and sealed by the enzyme ligase
In biochemistry, a ligase is an enzyme that can catalyze the joining ( ligation) of two large molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the larger molecules or the enz ...
.
Rate of replication
The rate of DNA replication in a living cell was first measured as the rate of phage T4 DNA elongation in phage-infected E. coli. During the period of exponential DNA increase at 37 °C, the rate was 749 nucleotides per second. The mutation rate per base pair per replication during phage T4 DNA synthesis is 1.7 per 108.Termination
Termination of DNA replication in ''E. coli'' is completed through the use of termination sequences and theTus protein
Tus, also known as terminus utilization substance, is a protein that binds to terminator sequences and acts as a counter-helicase when it comes in contact with an advancing helicase. The bound Tus protein effectively halts DNA polymerase movement. ...
. These sequences allow the two replication forks to pass through in only one direction, but not the other.
DNA replication initially produces two catenated or linked circular DNA duplexes, each comprising one parental strand and one newly synthesised strand (by nature of semiconservative replication
Semiconservative replication describe the mechanism of DNA replication in all known cells. DNA replication occurs on multiple origins of replication along the DNA template strand (antinsense strand). As the DNA double helix is unwound by helicase, ...
). This catenation can be visualised as two interlinked rings which cannot be separated. Topoisomerase 2 in ''E. coli'' unlinks or decatenates the two circular DNA duplexes by breaking the phosphodiester bonds present in two successive nucleotides of either parent DNA or newly formed DNA and thereafter the ligating activity ligates that broken DNA strand and so the two DNA get formed.
Other Prokaryotic replication models
The theta type replication has been already mentioned. There are other types of prokaryotic replication such asrolling circle replication
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA geno ...
and D-loop replication D-loop replication is a proposed process by which circular DNA like chloroplasts and mitochondria replicate their genetic material. An important component of understanding D-loop replication is that many chloroplasts and mitochondria have a single ...
Rolling Circle Replication
This is seen inbacterial conjugation
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteria ...
where the same circulartemplate DNA rotates and around it the new strand develops.
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecule ...
creates a nick
Nick may refer to:
* Nick (given name)
* A cricket term for a slight deviation of the ball off the edge of the bat
* British slang for being arrested
* British slang for a police station
* British slang for stealing
* Short for nickname
Place ...
in one of the strands of the conjugative plasmid at the ''oriT''. Relaxase may work alone or in a complex of over a dozen proteins known collectively as a relaxosome. In the F-plasmid system the relaxase enzyme is called TraI and the relaxosome consists of TraI, TraY, TraM and the integrated host factor IHF. The nicked strand, or ''T-strand'', is then unwound from the unbroken strand and transferred to the recipient cell in a 5'-terminus to 3'-terminus direction. The remaining strand is replicated either independent of conjugative action (vegetative replication beginning at the ''oriV'') or in concert with conjugation (conjugative replication similar to the rolling circle replication of lambda phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Leder ...
). Conjugative replication may require a second nick before successful transfer can occur. A recent report claims to have inhibited conjugation with chemicals that mimic an intermediate step of this second nicking event.
D-loop replication
D-loop replication is mostly seen in organellar DNA, Where a triple stranded structure calleddisplacement loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in this ca ...
is formed.
References
{{DNA replication DNA replication