Mariner-like Elements on:
[Wikipedia]
[Google]
[Amazon]
Tc1/mariner is a class and superfamily of
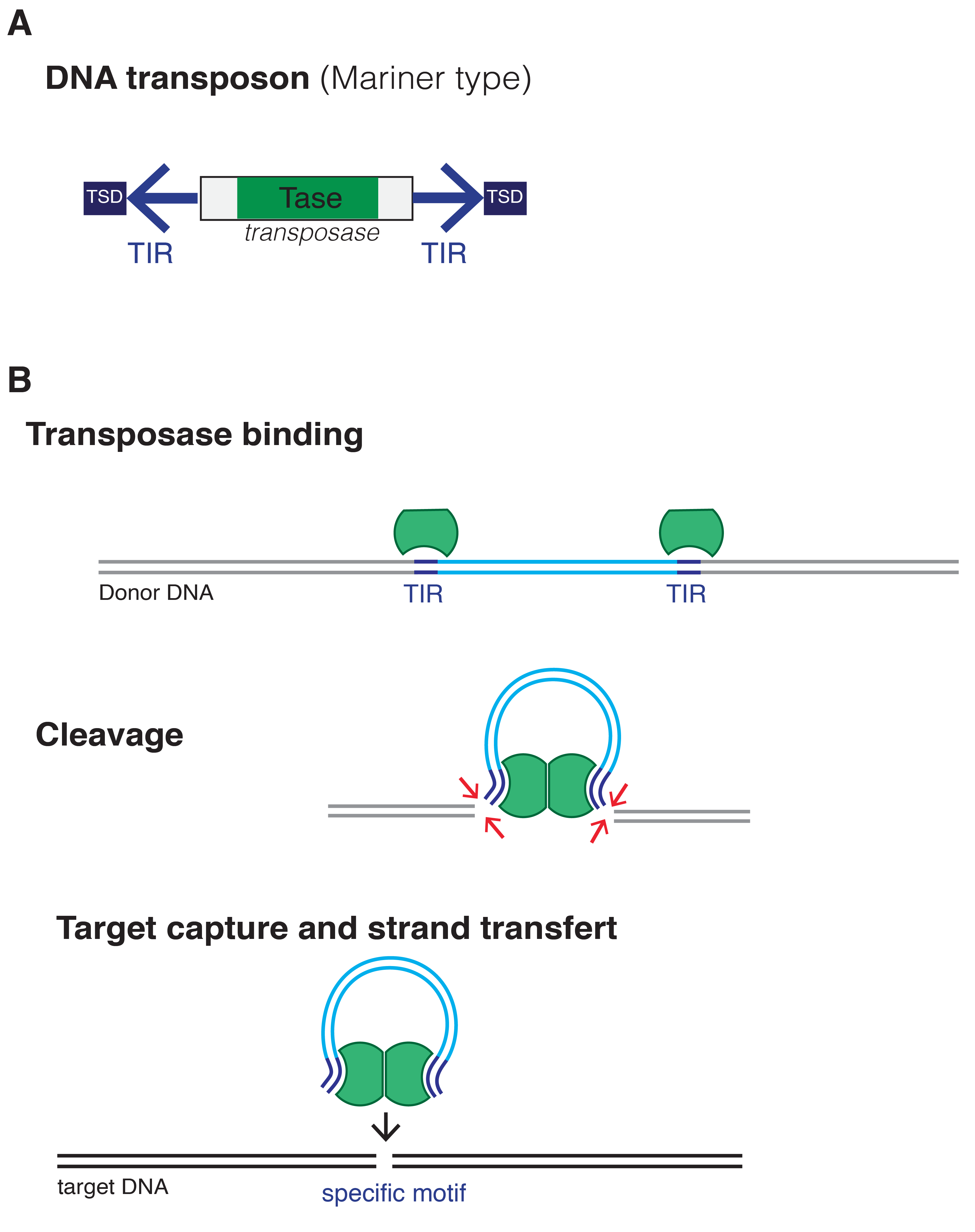 The transposon consists of a transposase gene, flanked by two terminal
The transposon consists of a transposase gene, flanked by two terminal
 The 360-amino acid polypeptide has three major subdomains: the amino-terminal DNA-recognition domain that is responsible for binding to the DR sequences in the mirrored IR/DR sequences of the transposon, a nuclear localization sequence (NLS), and a DDE domain that catalyzes the cut-and-paste set of reactions that comprise transposition. The DNA-recognition domain has two ''paired'' box sequences that can bind to DNA and are related to various motifs found on some transcription factors; the two ''paired'' boxes are labeled PAI and RED, both having the helix-turn-helix motif common for DNA-binding domains. The catalytic domain has the hallmark DDE (sometimes DDD) amino acids that are found in many transposase and recombinase enzymes. In addition, there is a region that is highly enriched in glycine (G) amino acids.
Several signatures for the superfamily of transcriptases have been given in various domain databases given the multi-domain nature of the protein. In addition, each domain are often represented by multiple entries, such as // among others for the "PAI" half of the box. The RED box is similarly dive rse (//, etc.) and is often in a winged HTH form for DNA recognition.
The 360-amino acid polypeptide has three major subdomains: the amino-terminal DNA-recognition domain that is responsible for binding to the DR sequences in the mirrored IR/DR sequences of the transposon, a nuclear localization sequence (NLS), and a DDE domain that catalyzes the cut-and-paste set of reactions that comprise transposition. The DNA-recognition domain has two ''paired'' box sequences that can bind to DNA and are related to various motifs found on some transcription factors; the two ''paired'' boxes are labeled PAI and RED, both having the helix-turn-helix motif common for DNA-binding domains. The catalytic domain has the hallmark DDE (sometimes DDD) amino acids that are found in many transposase and recombinase enzymes. In addition, there is a region that is highly enriched in glycine (G) amino acids.
Several signatures for the superfamily of transcriptases have been given in various domain databases given the multi-domain nature of the protein. In addition, each domain are often represented by multiple entries, such as // among others for the "PAI" half of the box. The RED box is similarly dive rse (//, etc.) and is often in a winged HTH form for DNA recognition.
interspersed repeat Interspersed repetitive DNA is found in all eukaryotic genomes. They differ from tandem repeat DNA in that rather than the repeat sequences coming right after one another, they are dispersed throughout the genome and nonadjacent. The sequence that r ...
s DNA (Class II) transposons. The elements of this class are found in all animals, including humans. They can also be found in protists and bacteria.
The class is named after its two best-studied members, the Tc1 transposon of ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (ro ...
'' and the mariner transposon of ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species ...
''.
Structure
 The transposon consists of a transposase gene, flanked by two terminal
The transposon consists of a transposase gene, flanked by two terminal inverted repeats An inverted repeat (or IR) is a single stranded sequence of nucleotides followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. ...
(TIR). Two short tandem site duplications (TSD) are present on both sides of the insert. Transposition happens when two transposases recognize and bind to TIR sequences, join together and promote DNA double-strand cleavage. The DNA-transposase complex then inserts its DNA cargo at specific DNA motifs elsewhere in the genome, creating short TSDs upon integration. In the IS630/Tc1/mariner system, the motif used is a "TA" dinucleotide, duplicated on both ends after insertion.
When the transposase gene is not carried by the transposon, it becomes a non-autonomous in that it now requires the gene to be expressed elsewhere to move around.
Transposase
 The 360-amino acid polypeptide has three major subdomains: the amino-terminal DNA-recognition domain that is responsible for binding to the DR sequences in the mirrored IR/DR sequences of the transposon, a nuclear localization sequence (NLS), and a DDE domain that catalyzes the cut-and-paste set of reactions that comprise transposition. The DNA-recognition domain has two ''paired'' box sequences that can bind to DNA and are related to various motifs found on some transcription factors; the two ''paired'' boxes are labeled PAI and RED, both having the helix-turn-helix motif common for DNA-binding domains. The catalytic domain has the hallmark DDE (sometimes DDD) amino acids that are found in many transposase and recombinase enzymes. In addition, there is a region that is highly enriched in glycine (G) amino acids.
Several signatures for the superfamily of transcriptases have been given in various domain databases given the multi-domain nature of the protein. In addition, each domain are often represented by multiple entries, such as // among others for the "PAI" half of the box. The RED box is similarly dive rse (//, etc.) and is often in a winged HTH form for DNA recognition.
The 360-amino acid polypeptide has three major subdomains: the amino-terminal DNA-recognition domain that is responsible for binding to the DR sequences in the mirrored IR/DR sequences of the transposon, a nuclear localization sequence (NLS), and a DDE domain that catalyzes the cut-and-paste set of reactions that comprise transposition. The DNA-recognition domain has two ''paired'' box sequences that can bind to DNA and are related to various motifs found on some transcription factors; the two ''paired'' boxes are labeled PAI and RED, both having the helix-turn-helix motif common for DNA-binding domains. The catalytic domain has the hallmark DDE (sometimes DDD) amino acids that are found in many transposase and recombinase enzymes. In addition, there is a region that is highly enriched in glycine (G) amino acids.
Several signatures for the superfamily of transcriptases have been given in various domain databases given the multi-domain nature of the protein. In addition, each domain are often represented by multiple entries, such as // among others for the "PAI" half of the box. The RED box is similarly dive rse (//, etc.) and is often in a winged HTH form for DNA recognition.
Sub-groups
The Tc1/mariner superfamily is generally subdivided by the catalytic domains of its transposase. It generally use a DDE (Asp-Asp-Glu) or DDD catalytic triad.Tc1
Tc1 (DD34E) is a transposon active in ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (ro ...
''. There are also Tc1-like transposons in humans, all inactive. Tc1-like elements are present in other lower vertebrates, including several fish species and amphibians.
In ''C. elegans'', it is a 1610 base-pair long sequence. Experiments show that this element "jumps" in human cells, with its transposase as the only protein required.
Another example of this family is Tc3, also a transposon found in ''C. elegans''.
Mariner
Mariner (DD34D) elements are found in multiple species, including humans. The Mariner transposon was first discovered by Jacobson and Hartl in ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species ...
'' in 1986. A classification of the group was published in 1993, which divided such sequences in insects into the mauritiana, cecropia, mellifera, irritans, and capitata subfamilies, after the types of insects they are found in. The classification does extend to other species.
This transposable element is known for its uncanny ability to be transmitted horizontally in many species. There are an estimated 14,000 copies of Mariner in the human genome comprising 2.6 million base pairs. The first mariner-element transposons outside of animal
Animals are multicellular, eukaryotic organisms in the Kingdom (biology), biological kingdom Animalia. With few exceptions, animals Heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, are Motilit ...
s were found in ''Trichomonas vaginalis
''Trichomonas vaginalis'' is an anaerobic, flagellated protozoan parasite and the causative agent of a sexually transmitted disease called trichomoniasis. It is the most common pathogenic protozoan that infects humans in industrialized countries ...
''.
Human Mariner-like transposons are divided into Hsmar1 (cecropia) and Hsmar2 (irritans) subfamilies. Although both types are inactive, one copy of Hsmar1 found in the SETMAR
Histone-lysine N-methyltransferase SETMAR is an enzyme that in humans is encoded by the ''SETMAR'' gene.
Function
SETMAR contains a SET domain that confers its histone methyltransferase activity, on Lys-4 and Lys-36 of Histone H3, both of which ...
gene is under selection as it provides DNA-binding for the histone-modifying protein. Hsmar2 has been reconstructed multiple times from the fossil sequences.
Mos1 (for ''Mosaic element'') was discovered in '' Drosophila mauritiana''. The Himar1 element has been isolated from the horn fly, ''Haematobia irritans
''Haematobia irritans'', the horn fly, is a small fly (about half the size of a common housefly). It was first described by Carl Linnaeus in his 1758 10th edition of ''Systema Naturae''. It is of the genus ''Haematobia'' which is the European ...
'' and can be used as a genetic tool in ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escher ...
''.
Other families
The rosa (DD41D) family is a family found in '' Ceratitis rosa''. Pogo/Fot1 (DDxD) is yet another family in this superfamily, x indicating a variable length. IS630, a mobile element in ''Shigella sonnei
''Shigella sonnei'' is a species of ''Shigella''. Together with ''Shigella flexneri'', it is responsible for 90% of shigellosis cases. ''Shigella sonnei'' is named for the Danish bacteriologist Carl Olaf Sonne. It is a Gram-negative, rod-shaped ...
'', also belongs to this superfamily.
A few additional new families with different lengths between the triads have been reported.
Pogo, also known as ''Tigger'' in humans, has been domesticated by humans and yeast alike into the CENPB
Centromere protein B also known as major centromere autoantigen B is an autoantigen protein of the cell nucleus. In humans, centromere protein B is encoded by the CENPB gene.
Function
Centromere protein B is a highly conserved protein that fa ...
gene. Other human domestications of pogo include TIGD1, TIGD2, TIGD3, TIGD4, TIGD5, TIGD6, TIGD7, JRK, JRKL, POGK, and POGZ
Pogo transposable element with ZNF domain is a protein that in humans is encoded by the ''POGZ'' gene.
The protein encoded by this gene appears to be a zinc finger protein containing a transposase domain at the C-terminus.
This protein was f ...
.
See also
*Sleeping Beauty transposon system
The ''Sleeping Beauty'' transposon system is a synthetic DNA transposon designed to introduce precisely defined DNA sequences into the chromosomes of vertebrate animals for the purposes of introducing new traits and to discover new genes and thei ...
* PiggyBac transposon system
References
{{DEFAULTSORT:Tc1 mariner DNA mobile genetic elements Human genetics Drosophila melanogaster genetics Caenorhabditis elegans genetics