DNA mismatch repair on:
[Wikipedia]
[Google]
[Amazon]
 DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during
DNA Repair
* {{DEFAULTSORT:Dna Mismatch Repair DNA repair
 DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
and recombination, as well as repairing
The technical meaning of maintenance involves functional checks, servicing, repairing or replacing of necessary devices, equipment, machinery, building infrastructure, and supporting utilities in industrial, business, and residential install ...
some forms of DNA damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
.
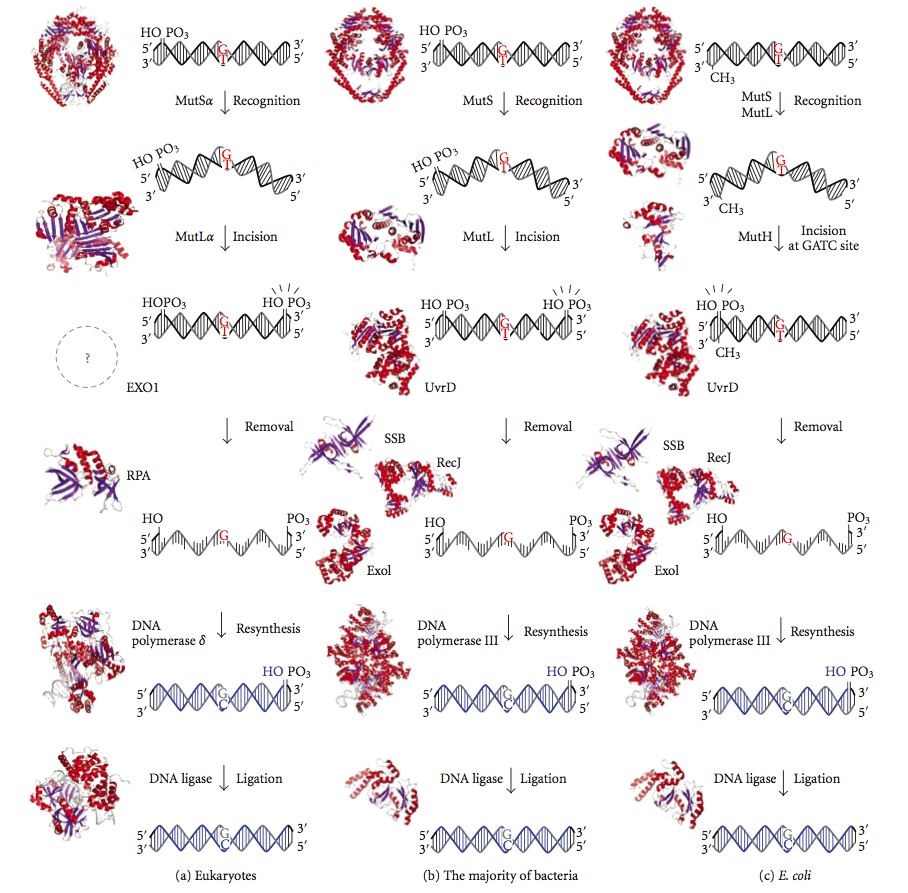
Mismatch repair is strand-specific. During DNA synthesis the newly synthesised (daughter) strand will commonly include errors. In order to begin repair, the mismatch repair machinery distinguishes the newly synthesised strand from the template (parental). In gram-negative bacteria, transient hemimethylation distinguishes the strands (the parental is methylated and daughter is not). However, in other prokaryotes and eukaryotes, the exact mechanism is not clear. It is suspected that, in eukaryotes, newly synthesized lagging-strand DNA transiently contains nicks
Nix (or Nicks) is a surname of English origin, which initially indicated that the person so named was the child of a person named Nicholas, traditionally shortened to "Nick". It is therefore closely related to Nixon and Nickson, which are derive ...
(before being sealed by DNA ligase) and provides a signal that directs mismatch proofreading systems to the appropriate strand. This implies that these nicks must be present in the leading strand, and evidence for this has recently been found.
Recent work has shown that nicks are sites for RFC-dependent loading of the replication sliding clamp, proliferating cell nuclear antigen (PCNA), in an orientation-specific manner, such that one face of the donut-shape protein is juxtaposed toward the 3'-OH end at the nick. Loaded PCNA then directs the action of the MutLalpha endonuclease to the daughter strand in the presence of a mismatch and MutSalpha or MutSbeta.
Any mutational event that disrupts the superhelical structure of DNA carries with it the potential to compromise the genetic stability of a cell. The fact that the damage detection and repair systems are as complex as the replication machinery itself highlights the importance evolution has attached to DNA fidelity.
Examples of mismatched bases include a G/T or A/C pairing (see DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
). Mismatches are commonly due to tautomerization
Tautomers () are structural isomers (constitutional isomers) of chemical compounds that readily interconvert.
The chemical reaction interconverting the two is called tautomerization. This conversion commonly results from the relocation of a hyd ...
of bases during DNA replication. The damage is repaired by recognition of the deformity caused by the mismatch, determining the template and non-template strand, and excising the wrongly incorporated base and replacing it with the correct nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecu ...
. The removal process involves more than just the mismatched nucleotide itself. A few or up to thousands of base pairs of the newly synthesized DNA strand can be removed.
Mismatch repair proteins
Mismatch repair is a highly conserved process fromprokaryotes
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
to eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
. The first evidence for mismatch repair was obtained from ''S. pneumoniae
''Streptococcus pneumoniae'', or pneumococcus, is a Gram-positive, spherical bacteria, alpha-hemolytic (under aerobic conditions) or beta-hemolytic (under anaerobic conditions), aerotolerant anaerobic member of the genus Streptococcus. They are ...
'' (the hexA and hexB gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s). Subsequent work on '' E. coli'' has identified a number of genes that, when mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
ally inactivated, cause hypermutable strains. The gene products are, therefore, called the "Mut" proteins, and are the major active components of the mismatch repair system. Three of these proteins are essential in detecting the mismatch and directing repair machinery to it: MutS, MutH and MutL (MutS is a homologue of HexA and MutL of HexB).
MutS forms a dimer (MutS2) that recognises the mismatched base on the daughter strand and binds the mutated DNA. MutH binds at hemimethylated sites along the daughter DNA, but its action is latent, being activated only upon contact by a MutL dimer (MutL2), which binds the MutS-DNA complex and acts as a mediator between MutS2 and MutH, activating the latter. The DNA is looped out to search for the nearest d(GATC) methylation site to the mismatch, which could be up to 1 kb away. Upon activation by the MutS-DNA complex, MutH nicks the daughter strand near the hemimethylated site. MutL recruits UvrD helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separatin ...
(DNA Helicase II) to separate the two strands with a specific 3' to 5' polarity. The entire MutSHL complex then slides along the DNA in the direction of the mismatch, liberating the strand to be excised as it goes. An exonuclease trails the complex and digests the ss-DNA tail. The exonuclease recruited is dependent on which side of the mismatch MutH incises the strand – 5' or 3'. If the nick made by MutH is on the 5' end of the mismatch, either RecJ or ExoVII (both 5' to 3' exonucleases) is used. If, however, the nick is on the 3' end of the mismatch, ExoI (a 3' to 5' enzyme) is used.
The entire process ends past the mismatch site - i.e., both the site itself and its surrounding nucleotides are fully excised. The single-strand gap created by the exonuclease can then be repaired by DNA Polymerase III (assisted by single-strand-binding protein), which uses the other strand as a template, and finally sealed by DNA ligase. DNA methylase then rapidly methylates the daughter strand.
MutS homologs
When bound, the MutS2 dimer bends the DNA helix and shields approximately 20 base pairs. It has weak ATPase activity, and binding of ATP leads to the formation of tertiary structures on the surface of the molecule. Thecrystal structure
In crystallography, crystal structure is a description of the ordered arrangement of atoms, ions or molecules in a crystalline material. Ordered structures occur from the intrinsic nature of the constituent particles to form symmetric pattern ...
of MutS reveals that it is exceptionally asymmetric, and, while its active conformation is a dimer, only one of the two halves interacts with the mismatch site.
In eukaryotes, MutS homologs form two major heterodimers: Msh2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that cod ...
/Msh6 (MutSα) and Msh2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that cod ...
/Msh3 (MutSβ). The MutSα pathway is involved primarily in base substitution and small-loop mismatch repair. The MutSβ pathway is also involved in small-loop repair, in addition to large-loop (~10 nucleotide loops) repair. However, MutSβ does not repair base substitutions.
MutL homologs
MutL also has weak ATPase activity (it uses ATP for purposes of movement). It forms a complex with MutS and MutH, increasing the MutS footprint on the DNA. However, the processivity (the distance the enzyme can move along the DNA before dissociating) of UvrD is only ~40–50 bp. Because the distance between the nick created by MutH and the mismatch can average ~600 bp, if there is not another UvrD loaded the unwound section is then free to re-anneal to its complementary strand, forcing the process to start over. However, when assisted by MutL, the ''rate'' of UvrD loading is greatly increased. While the processivity (and ATP utilisation) of the individual UvrD molecules remains the same, the total effect on the DNA is boosted considerably; the DNA has no chance to re-anneal, as each UvrD unwinds 40-50 bp of DNA, dissociates, and then is immediately replaced by another UvrD, repeating the process. This exposes large sections of DNA toexonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is ...
digestion, allowing for quick excision (and later replacement) of the incorrect DNA.
Eukaryotes have five MutL homologs designated as MLH1, MLH2, MLH3, PMS1, and PMS2. They form heterodimers that mimic MutL in ''E. coli''. The human homologs of prokaryotic MutL form three complexes referred to as MutLα, MutLβ, and MutLγ. The MutLα complex is made of MLH1 and PMS2 subunits, the MutLβ heterodimer is made of MLH1 and PMS1, whereas MutLγ is made of MLH1 and MLH3. MutLα acts as an endonuclease that introduces strand breaks in the daughter strand upon activation by mismatch and other required proteins, MutSα and PCNA. These strand interruptions serve as entry points for an exonuclease activity that removes mismatched DNA. Roles played by MutLβ and MutLγ in mismatch repair are less-understood.
MutH: an endonuclease present in ''E. coli'' and ''Salmonella''
MutH is a very weakendonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonuclease ...
that is activated once bound to MutL (which itself is bound to MutS). It nicks unmethylated DNA and the unmethylated strand of hemimethylated DNA but does not nick fully methylated DNA. Experiments have shown that mismatch repair is random if neither strand is methylated. These behaviours led to the proposal that MutH determines which strand contains the mismatch.
MutH has no eukaryotic homolog. Its endonuclease function is taken up by MutL homologs, which have some specialized 5'-3' exonuclease activity. The strand bias for removing mismatches from the newly synthesized daughter strand in eukaryotes may be provided by the free 3' ends of Okazaki fragments in the new strand created during replication.
PCNA β-sliding clamp
PCNA and the β-sliding clamp associate with MutSα/β and MutS, respectively. Although initial reports suggested that the PCNA-MutSα complex may enhance mismatch recognition, it has been recently demonstrated that there is no apparent change in affinity of MutSα for a mismatch in the presence or absence of PCNA. Furthermore, mutants of MutSα that are unable to interact with PCNA ''in vitro
''In vitro'' (meaning in glass, or ''in the glass'') studies are performed with microorganisms, cells, or biological molecules outside their normal biological context. Colloquially called " test-tube experiments", these studies in biology a ...
'' exhibit the capacity to carry out mismatch recognition and mismatch excision to near wild type levels. Such mutants are defective in the repair reaction directed by a 5' strand break, suggesting for the first time MutSα function in a post-excision step of the reaction.
Clinical significance
Inherited defects in mismatch repair
Mutations in the human homologues of the Mut proteins affect genomic stability, which can result inmicrosatellite instability
Microsatellite instability (MSI) is the condition of genetic hypermutability (predisposition to mutation) that results from impaired DNA mismatch repair (MMR). The presence of MSI represents phenotypic evidence that MMR is not functioning norma ...
(MSI), implicated in some human cancers. In specific, the hereditary nonpolyposis colorectal cancers ( HNPCC or Lynch syndrome) are attributed to damaging germline variants in the genes encoding the MutS and MutL homologues MSH2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that cod ...
and MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH ...
respectively, which are thus classified as tumour suppressor genes. One subtype of HNPCC, the Muir-Torre Syndrome (MTS), is associated with skin tumors. If both inherited copies (alleles) of a MMR gene bear damaging genetic variants, this results in a very rare and severe condition: the mismatch repair cancer syndrome (or constitutional mismatch repair deficiency, CMMR-D), manifesting as multiple occurrences of tumors at an early age, often colon and brain tumor
A brain tumor occurs when abnormal cells form within the brain. There are two main types of tumors: malignant tumors and benign (non-cancerous) tumors. These can be further classified as primary tumors, which start within the brain, and seco ...
s.
Epigenetic silencing of mismatch repair genes
Sporadic cancers with a DNA repair deficiency only rarely have a mutation in a DNA repair gene, but they instead tend to haveepigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are ...
alterations such as promoter methylation that inhibit DNA repair gene expression. About 13% of colorectal cancers are deficient in DNA mismatch repair, commonly due to loss of MLH1 (9.8%), or sometimes MSH2, MSH6 or PMS2 (all ≤1.5%). For most MLH1-deficient sporadic colorectal cancers, the deficiency was due to MLH1 promoter methylation. Other cancer types have higher frequencies of MLH1 loss (see table below), which are again largely a result of methylation of the promoter of the ''MLH1'' gene. A different epigenetic mechanism underlying MMR deficiencies might involve over-expression of a microRNA, for example miR-155
MiR-155 is a microRNA that in humans is encoded by the ''MIR155'' host gene or ''MIR155HG''. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control '' in vivo'' of miR-155 expression may inhibit mal ...
levels inversely correlate with expression of MLH1 or MSH2 in colorectal cancer.
MMR failures in field defects
A field defect (field cancerization) is an area of epithelium that has been preconditioned by epigenetic or genetic changes, predisposing it towards development of cancer. As pointed out by Rubin " ...there is evidence that more than 80% of the somatic mutations found in mutator phenotype human colorectal tumors occur before the onset of terminal clonal expansion." Similarly, Vogelstein et al. point out that more than half of somatic mutations identified in tumors occurred in a pre-neoplastic phase (in a field defect), during growth of apparently normal cells. MLH1 deficiencies were common in the field defects (histologically normal tissues) surrounding tumors; see Table above. Epigenetically silenced or mutated MLH1 would likely not confer a selective advantage upon a stem cell, however, it would cause increased mutation rates, and one or more of the mutated genes may provide the cell with a selective advantage. The deficient''MLH1'' gene could then be carried along as a selectively near-neutral passenger (hitch-hiker) gene when the mutated stem cell generates an expanded clone. The continued presence of a clone with an epigenetically repressed ''MLH1'' would continue to generate further mutations, some of which could produce a tumor.MMR components in humans
In humans, seven DNA mismatch repair (MMR) proteins (MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH ...
, MLH3
DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the ''MLH3'' gene.
Function
This gene is a member of the MutL-homolog (MLH) family of DNA mismatch repair (MMR) genes. MLH genes are implicated in maintaining genomic i ...
, MSH2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that cod ...
, MSH3, MSH6, PMS1 and PMS2
Mismatch repair endonuclease PMS2 is an enzyme that in humans is encoded by the ''PMS2'' gene.
Function
This gene is one of the PMS2 gene family members which are found in clusters on chromosome 7. Human PMS2 related genes are located at band ...
) work coordinately in sequential steps to initiate repair of DNA mismatches. In addition, there are Exo1-dependent and Exo1-independent MMR subpathways.
Other gene products involved in mismatch repair (subsequent to initiation by MMR genes) in humans include DNA polymerase delta, PCNA, RPA, HMGB1, RFC and DNA ligase I, plus histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
and chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
modifying factors.
In certain circumstances, the MMR pathway may recruit an error-prone DNA polymerase eta ( POLH). This happens in B-lymphocytes during somatic hypermutation
Somatic hypermutation (or SHM) is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it (e.g. microbes), as seen during class switching. A major component of the process of affinity maturation, SHM ...
, where POLH is used to introduce genetic variation into antibody genes. However, this error-prone MMR pathway may be triggered in other types of human cells upon exposure to genotoxins and indeed it is broadly active in various human cancers, causing mutations that bear a signature of POLH activity.
MMR and mutation frequency
Recognizing and repairing mismatches and indels is important for cells because failure to do so results inmicrosatellite instability
Microsatellite instability (MSI) is the condition of genetic hypermutability (predisposition to mutation) that results from impaired DNA mismatch repair (MMR). The presence of MSI represents phenotypic evidence that MMR is not functioning norma ...
(MSI) and an elevated spontaneous mutation rate
In genetics, the mutation rate is the frequency of new mutations in a single gene or organism over time. Mutation rates are not constant and are not limited to a single type of mutation; there are many different types of mutations. Mutation rates ...
(mutator phenotype). In comparison to other cancer types, MMR-deficient (MSI) cancer has a very high frequency of mutations, close to melanoma and lung cancer, cancer types caused by much exposure to UV radiation and mutagenic chemicals.
In addition to a very high mutation burden, MMR deficiencies result in an unusual distribution of somatic mutations across the human genome: this suggests that MMR preferentially protects the gene-rich, early-replicating euchromatic regions. In contrast, the gene-poor, late-replicating heterochromatic genome regions exhibit high mutation rates in many human tumors.
The histone modification H3K36me3
H3K36me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri- methylation at the 36th lysine residue of the histone H3 protein and often associated with gene bodies.
There are diverse modif ...
, an epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are ...
mark of active chromatin, has the ability to recruit the MSH2-MSH6 (hMutSα) complex. Consistently, regions of the human genome with high levels of H3K36me3 accumulate less mutations due to MMR activity.
Loss of multiple DNA repair pathways in tumors
Lack of MMR often occurs in coordination with loss of other DNA repair genes. For example, MMR genes ''MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH ...
and'' ''MLH3
DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the ''MLH3'' gene.
Function
This gene is a member of the MutL-homolog (MLH) family of DNA mismatch repair (MMR) genes. MLH genes are implicated in maintaining genomic i ...
'' as well as 11 other DNA repair genes (such as ''MGMT
MGMT () is an American indie rock band formed in 2002 in Middletown, Connecticut. It was founded by multi-instrumentalists Andrew VanWyngarden and Ben Goldwasser. Alongside VanWyngarden and Goldwasser, MGMT's live lineup currently consists o ...
'' and many NER pathway genes) were significantly down-regulated in lower grade as well as in higher grade astrocytomas, in contrast to normal brain tissue. Moreover, MLH1 and MGMT
MGMT () is an American indie rock band formed in 2002 in Middletown, Connecticut. It was founded by multi-instrumentalists Andrew VanWyngarden and Ben Goldwasser. Alongside VanWyngarden and Goldwasser, MGMT's live lineup currently consists o ...
expression was closely correlated in 135 specimens of gastric cancer and loss of MLH1 and MGMT appeared to be synchronously accelerated during tumor progression.
Deficient expression of multiple DNA repair genes is often found in cancers, and may contribute to the thousands of mutations usually found in cancers (see Mutation frequencies in cancers).
Aging
A popular idea, that has failed to gain significant experimental support, is the idea that mutation, as distinct from DNA damage, is the primary cause of aging. Mice defective in the ''mutL'' homolog ''Pms2'' have about a 100-fold elevated mutation frequency in all tissues, but do not appear to age more rapidly. These mice display mostly normal development and life, except for early onset carcinogenesis and male infertility.See also
*Base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from ...
*Nucleotide excision repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucle ...
References
Further reading
* * * * * * *External links
DNA Repair
* {{DEFAULTSORT:Dna Mismatch Repair DNA repair