DNA Shuffling on:
[Wikipedia]
[Google]
[Amazon]
 DNA shuffling, also known as molecular breeding, is an
DNA shuffling, also known as molecular breeding, is an
 DNA shuffling, also known as molecular breeding, is an
DNA shuffling, also known as molecular breeding, is an in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
random recombination method to generate mutant genes
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
for directed evolution and to enable a rapid increase in DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
library size. Three procedures for accomplishing DNA shuffling are molecular breeding which relies on homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
or the similarity of the DNA sequences, restriction enzymes which rely on common restriction sites, and nonhomologous random recombination which requires the use of hairpins. In all of these techniques, the parent genes are fragmented and then recombined.
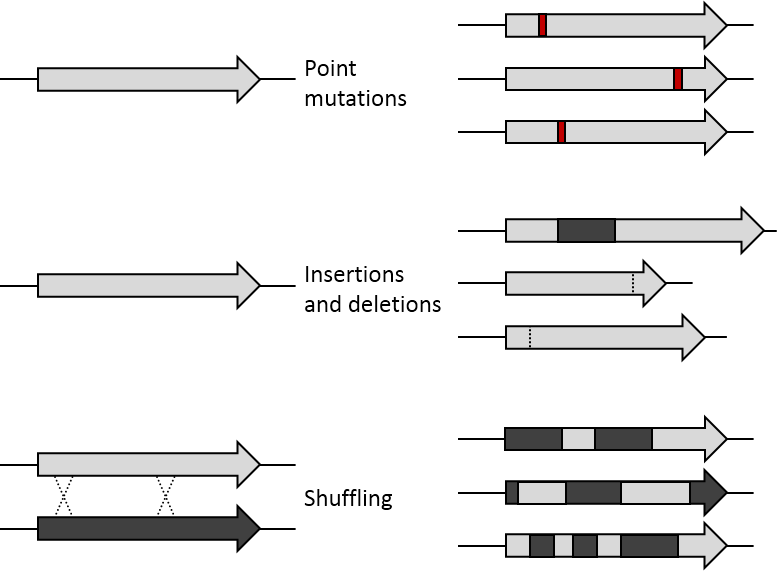
DNA shuffling utilizes random recombination as opposed to site-directed mutagenesis in order to generate proteins with unique attributes or combinations of desirable characteristics encoded in the parent genes such as thermostability and high activity. The potential for DNA shuffling to produce novel proteins is exemplified by the figure shown on the right which demonstrates the difference between point mutations, insertions and deletions, and DNA shuffling. Specifically, this figure shows the use of DNA shuffling on two parent genes which enables the generation of recombinant proteins that have a random combination of sequences from each parent gene. This is distinct from point mutations in which one nucleotide has been changed, inserted, or deleted and insertions or deletions where a sequence of nucleotides has been added or removed, respectively. As a result of the random recombination, DNA shuffling is able to produce proteins with new qualities or multiple advantageous features derived from the parent genes.
In 1994, Willem P.C. Stemmer published the first paper on DNA shuffling. Since the introduction of the technique, DNA shuffling has been applied to protein and small molecule pharmaceuticals, bioremediation
Bioremediation broadly refers to any process wherein a biological system (typically bacteria, microalgae, fungi in mycoremediation, and plants in phytoremediation), living or dead, is employed for removing environmental pollutants from air, wate ...
, vaccines, gene therapy, and evolved viruses. Other techniques which yield similar results to DNA shuffling include random chimeragenesis on transient templates (RACHITT), random printing in vitro recombination (RPR), and the staggered extension process (StEP).
History
DNA shuffling by molecular breeding was first reported in 1994 by Willem P.C. Stemmer. He started by fragmenting the β-lactamase gene that had been amplified with thepolymerase chain reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed st ...
(PCR) by using DNase I, which randomly cleaves DNA. He then completed a modified PCR reaction where primers were not employed which resulted in the annealing of homologous fragments or fragments with similar sequences. Finally, these fragments were amplified by PCR. Stemmer reported that the use of DNA shuffling in combination with backcrossing resulted in the elimination of non-essential mutations
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosi ...
and an increase in the production of the antibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting pathogenic bacteria, bacterial infections, and antibiotic medications are widely used in the therapy ...
cefotaxime
Cefotaxime is an antibiotic used to treat several bacterial infections in humans, other animals, and plant tissue culture. Specifically in humans it is used to treat joint infections, pelvic inflammatory disease, meningitis, pneumonia, urin ...
. He also emphasized the potential for molecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
with DNA shuffling. Specifically, he indicated the technique could be used to modify proteins.
DNA shuffling has since been applied to generate libraries of hybrid or chimeric genes and has inspired family shuffling which is defined as the use of related genes in DNA shuffling. Additionally, DNA shuffling has been applied to protein and small molecule pharmaceuticals, bioremediation, gene therapy, vaccines, and evolved viruses.
Procedures
Molecular breeding
First, DNase I is used to fragment a set of parent genes into segments of double stranded DNA ranging from 10-50 bp to more than 1 kbp. This is followed by a PCR without primers. In the PCR, DNA fragments with sufficiently overlapping sequences will anneal to each other and then be extended byDNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
. The PCR extension will not occur unless there are DNA sequences of high similarity. The important factors influencing the sequences synthesized in DNA shuffling are the DNA polymerase, salt concentrations, and annealing temperature. For example, the use of Taq polymerase for amplification of a 1 kbp fragment in a PCR of 20 cycles results in 33% to 98% of the products containing one or more mutations.
Multiple cycles of PCR extension can be used to amplify the fragments. The addition of primers that are designed to be complementary to the ends of the extended fragments are added to further amplify the sequences with another PCR. Primers may be chosen to have additional sequences added on to their 5’ ends, such as sequences for restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
recognition sites which are needed for ligation into a cloning vector.
It is possible to recombine portions of the parent genes to generate hybrids or chimeric forms with unique properties, hence the term DNA shuffling. The disadvantage of molecular breeding is the requirement for the similarity between the sequences, which has inspired the development of other procedures for DNA shuffling.
Restriction enzymes
Restriction enzymes are employed to fragment the parent genes. The fragments are then joined together through ligation which can be accomplished with DNA ligase. For example, if two parent genes have three restriction sites fourteen different full-length gene hybrids can be created. The number of unique full-length hybrids is determined by the fact that a gene with three restriction sites can be broken up into four fragments. Thus, there are two options for each of the four positions minus the combinations that would recreate the two parent genes yielding 24 - 2 = 14 different full-length hybrid genes. The main difference between DNA shuffling with restriction enzymes and molecular breeding is molecular breeding relies on the homology of the sequences for the annealing of the strands and PCR for extension whereas by using restriction enzymes, fragment ends that can be ligated are created. The main advantages of using restriction enzymes include control over the number of recombination events and lack of PCR amplification requirement. The main disadvantage is the requirement of common restriction enzyme sites.Nonhomologous random recombination
In order to generate segments ranging from 10-50 bp to more than 1 kb, DNase I is utilized. The ends of the fragments are made blunt by adding T4 DNA polymerase. Blunting the fragments is important for combining the fragments as incompatible sticky-ends, or overhangs, prevent end joining. Hairpins with a specific restriction site are then added to the mixture of fragments. Next, T4 DNA ligase is employed to ligate the fragments to form extended sequences. The ligation of the hairpins to the fragments limits the length of the extended sequences by preventing the addition of more fragments. Finally, in order to remove the hairpin loops, a restriction enzyme is utilized. Nonhomologous random recombination differs from molecular breeding as homology of the ligated sequences is not necessary which is an advantage. However, because this process recombines the fragments randomly it is probable that a large fraction of the recombined DNA sequences will not have the desired characteristics which is a disadvantage. Nonhomologous random recombination also differs from the use of restriction enzymes for DNA shuffling as common restriction enzyme sites on the parent genes are not required and the use of hairpins is necessary which demonstrates an advantage and disadvantage of nonhomologous random recombination over the use of restriction enzymes, respectively.Applications
Protein and small molecule pharmaceuticals
Since DNA shuffling enables the recombination of genes, protein activities can be enhanced. For example, DNA shuffling has been used to increase the potency of phage-displayed recombinantinterferons
Interferons (IFNs, ) are a group of signaling proteins made and released by host cells in response to the presence of several viruses. In a typical scenario, a virus-infected cell will release interferons causing nearby cell (biology), cell ...
on murine and human cells. Additionally, the improvement of green fluorescent protein (GFP) was accomplished with DNA shuffling by molecular breeding as a 45-fold greater signal than the standard for whole cell fluorescence was obtained. Furthermore, the synthesis of diverse genes can also result in the production of proteins with novel attributes. Therefore, DNA shuffling has been used to develop proteins to detoxify chemicals. For example, the homologous recombination method of DNA shuffling by molecular breeding has been utilized to enhance the detoxification of atrazine
Atrazine ( ) is a Organochlorine compound, chlorinated herbicide of the triazine class. It is used to prevent pre-emergence broadleaf weeds in crops such as maize (corn), soybean and sugarcane and on turf, such as golf courses and residential law ...
and arsenate.
Bioremediation
DNA shuffling has also been used to improve the degradation of biological pollutants. Specifically, a recombinant E. coli strain has been created with the use of DNA shuffling by molecular breeding for the bioremediation of trichloroethylene (TCE), a potentialcarcinogen
A carcinogen () is any agent that promotes the development of cancer. Carcinogens can include synthetic chemicals, naturally occurring substances, physical agents such as ionizing and non-ionizing radiation, and biologic agents such as viruse ...
, which is less susceptible to toxic epoxide intermediates.
Vaccines
The ability to select desirable recombinants with DNA shuffling has been used in combination with screening strategies to enhance vaccine candidates against infections with an emphasis on improving immunogenicity, vaccine production, stability, and cross-reactivity to multiple strains of pathogens. Some vaccine candidates forPlasmodium falciparum
''Plasmodium falciparum'' is a Unicellular organism, unicellular protozoan parasite of humans and is the deadliest species of ''Plasmodium'' that causes malaria in humans. The parasite is transmitted through the bite of a female ''Anopheles'' mos ...
, dengue virus, encephalitic alphaviruses (including: VEEV, WEEV, and EEEV), human immunodeficiency virus-1 (HIV-1), and hepatitis B virus (HBV) have been investigated.
Gene therapy and evolved viruses
The requirements for human gene therapies include high purity, high- titer, and stability. DNA shuffling allows for the fabrication of retroviral vectors with these attributes. For example, DNA shuffling with molecular breeding was applied to six ecotropic murine leukemia virus (MLV) strains which resulted in the compilation of an extensive library of recombinant retrovirus and the identification of multiple clones with increased stability. Furthermore, the application of DNA shuffling by molecular breeding on multiple parent adeno-associated virus (AAV) vectors was employed to generate a library of ten million chimeras. The advantageous attributes obtained include increased resistance to human intravenous immunoglobulin (IVIG) and the production of cell tropism in the novel viruses.Comparison to other techniques
While DNA shuffling has become a useful technique for random recombination, other methods including RACHITT, RPR, and StEP have also been developed for this purpose. Below are some advantages and disadvantages of these other methods for recombination.RACHITT
In RACHITT, fragments of single stranded (ss) parent genes are annealed onto a ss template resulting in decreased mismatching which is an advantage. Additionally, RACHIIT enables genes with low sequence similarity to be recombined. However, a major disadvantage is the preparation of the ss fragments of the parent genes and ss template.RPR
RPR makes use of random primers. These random primers are annealed to template DNA and are then extended by the Klenow fragment. Next, the templates are removed and the fragments are assembled by homology in a process similar to PCR. Some major benefits include the smaller requirement for parent genes due to the use of ss templates and increased sequence diversity by mispriming and misincorporation. One disadvantage of RPR is the preparation of the template.StEP
In StEP, brief cycles of primer annealing to a template and extension by polymerase are employed to generate full-length sequences.{{cite book , vauthors = Aguinaldo AM, Arnold FH , title = Directed Evolution Library Creation , chapter = Staggered extension process (StEP) in vitro recombination , volume = 231 , pages = 105–110 , date = 2003 , pmid = 12824608 , doi = 10.1385/1-59259-395-X:105 , publisher = Humana Press , isbn = 978-1-59259-395-8 , series = Methods in Molecular Biology , veditors = Arnold FH, Georgiou G , place = Totowa, NJ The main advantages of StEP are the simplicity of the method and the lack of fragment purification. The disadvantages of StEP include that it is time consuming and requires sequence homology.See also
* SCOPE (protein engineering)References