|
Untranslated Regions
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs. Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides. The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translation. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important list of RNAs, types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, Extracellular RNA, exRNAs, scaRNAs and the long noncoding RNA, long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent Transcriptomics, transcriptomic and Bioinformatics, bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kozak Consensus Sequence
The Kozak consensus sequence (Kozak consensus or Kozak sequence) is a nucleic acid motif that functions as the protein translation initiation site in most eukaryotic mRNA transcripts. Regarded as the optimum sequence for initiating translation in eukaryotes, the sequence is an integral aspect of protein regulation and overall cellular health as well as having implications in human disease. It ensures that a protein is correctly translated from the genetic message, mediating ribosome assembly and translation initiation. A wrong start site can result in non-functional proteins. As it has become more studied, expansions of the nucleotide sequence, bases of importance, and notable exceptions have arisen. The sequence was named after the scientist who discovered it, Marilyn Kozak. Kozak discovered the sequence through a detailed analysis of DNA genomic sequences. The Kozak sequence is not to be confused with the ribosomal binding site (RBS), that being either the 5′ cap of a m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Globin
The globins are a superfamily of heme-containing globular proteins, involved in binding and/or transporting oxygen. These proteins all incorporate the globin fold, a series of eight alpha helical segments. Two prominent members include myoglobin and hemoglobin. Both of these proteins reversibly bind oxygen via a heme prosthetic group. They are widely distributed in many organisms. Structure Globin superfamily members share a common three-dimensional fold. This 'globin fold' typically consists of eight alpha helices, although some proteins have additional helix extensions at their termini. Since the globin fold contains only helices, it is classified as an all-alpha protein fold. The globin fold is found in its namesake globin families as well as in phycocyanins. The globin fold was thus the first protein fold discovered (myoglobin was the first protein whose structure was solved). Helix packaging The eight helices of the globin fold core share significant nonlocal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
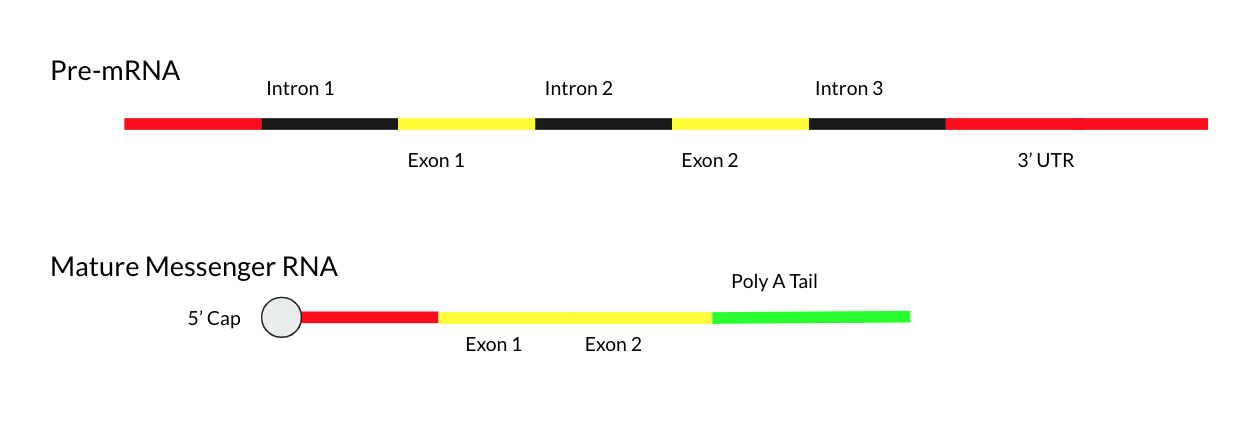
Mature Messenger RNA
Mature messenger RNA, often abbreviated as mature mRNA is a eukaryotic RNA transcript that has been spliced and processed and is ready for translation in the course of protein synthesis. Unlike the eukaryotic Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ... RNA immediately after transcription known as precursor messenger RNA, mature mRNA consists exclusively of exons and has all introns removed. Mature mRNA is also called "mature transcript", "mature RNA" or "mRNA". The production of a mature mRNA molecule occurs in 3 steps: # Capping of the 5' end # Polyadenylation of the 3' end # RNA Splicing of the introns Capping the 5' End During capping, a 7-methylguanosine residue is attached to the 5'-terminal end of the primary transcripts.This is otherwise known as the GTP ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear genes, nuclear-encoded genes, splicing occurs in the cell nucleus, nucleus either during or immediately after Transcription (biology), transcription. For those eukaryotic transcription, eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translation (biology), translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron [i.e., gene] ... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA Transcription (genetics), transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primary Transcript
A primary transcript is the single-stranded ribonucleic acid ( RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing. Pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely processed, it is termed "mature messenger RNA", or simply "messenger RNA". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA. There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important list of RNAs, types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, Extracellular RNA, exRNAs, scaRNAs and the long noncoding RNA, long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent Transcriptomics, transcriptomic and Bioinformatics, bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |