|
Ubiquitylation
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. Proteins are tagged for degradation with a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a ''polyubiquitin chain'' that is bound by the proteasome, allowing it to degrade the tagged protein. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteasomes are found inside all eukaryotes and archaea, and in some ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin C
Polyubiquitin-C is a protein encoded by the ''UBC'' gene in humans. Polyubiquitin-C is one of the sources of ubiquitin, along with UBB, UBA52, and RPS27A. ''UBC'' gene is one of the two stress-regulated polyubiquitin genes (''UBB'' and ''UBC'') in mammals. It plays a key role in maintaining cellular ubiquitin levels under stress conditions. Defects of ''UBC'' gene could lead to mid-gestation embryonic lethality. Structure Gene ''UBC'' gene is located at chromosome 12q24.3, consisting of 2 exons. The promoter of the ''UBC'' gene contains putative heat shock elements ( HSEs), which mediates UBC induction upon stress. ''UBC'' gene differs from ''UBB'' gene in the number of Ub coding units they contain. Nine to ten Ub units were in the ''UBC'' gene. Protein In polyubiquitin-C, the C-terminus of a given ubiquitin molecule is covalently conjugated to either the N-terminal residue or one of seven lysine residues of another ubiquitin molecule. Different linking of ubiquitin ch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-conjugating Enzyme
Ubiquitin-conjugating enzymes, also known as E2 enzymes and more rarely as ''ubiquitin-carrier enzymes'', perform the second step in the ubiquitination reaction that targets a protein for degradation via the proteasome. The ubiquitination process covalently attaches ubiquitin, a short protein of 76 amino acids, to a lysine residue on the target protein. Once a protein has been tagged with one ubiquitin molecule, additional rounds of ubiquitination form a polyubiquitin chain that is recognized by the proteasome's 19S regulatory particle, triggering the ATP-dependent unfolding of the target protein that allows passage into the proteasome's 20S core particle, where proteases degrade the target into short peptide fragments for recycling by the cell. Relationships A ubiquitin-activating enzyme, or E1, first activates the ubiquitin by covalently attaching the molecule to its active site cysteine residue. The activated ubiquitin is then transferred to an E2 cysteine. Once conjugated t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-activating Enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which (among other things) can target a protein for degradation via a proteasome. This covalent bond of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms. Many processes such as cell division, immune responses and embryonic development are also regulated by post-translational modification by ubiquitin and ubiquitin-like proteins. Ubiquitination (ubiquitylation) Ubiquitin-activating enzyme (E1) starts the ubiquitination process (Figure 1). The E1 enzyme, along with ATP, binds to the ubiquitin protein. The E1 enzyme then passes the ubiquitin protein to a second protein, called ubiquitin carrier or conjugation protein (E2). The E2 protein complexes with a ubiquitin protein ligase (E3). This ubiquitin protein ligase recognizes which protein needs to be tagged and catalyzes the transfer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Atomic Mass Unit
The dalton or unified atomic mass unit (symbols: Da or u) is a non-SI unit of mass widely used in physics and chemistry. It is defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. The atomic mass constant, denoted ''m''u, is defined identically, giving . This unit is commonly used in physics and chemistry to express the mass of atomic-scale objects, such as atoms, molecules, and elementary particles, both for discrete instances and multiple types of ensemble averages. For example, an atom of helium-4 has a mass of . This is an intrinsic property of the isotope and all helium-4 atoms have the same mass. Acetylsalicylic acid (aspirin), , has an average mass of approximately . However, there are no acetylsalicylic acid molecules with this mass. The two most common masses of individual acetylsalicylic acid molecules are , having the most common isotopes, and , in which one carbon is carbon-13. The molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), a carboxyl group (which is in the deprotonated −COO− form under biological conditions), and a side chain containing a hydroxyl group, making it a polar, uncharged amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Threonine is synthesized from aspartate in bacteria such as ''E. coli''. It is encoded by all the codons starting AC (ACU, ACC, ACA, and ACG). Threonine sidechains are often hydrogen bonded; the most common small motifs formed are based on interactions with serine: ST turns, ST motifs (often at the beginning of alpha helices) and ST staples (usually at the middle of alpha helices). Modifications The threonine residue is susceptible to numerous posttranslational modifications. The hydroxyl side-chain can underg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ester
In chemistry, an ester is a compound derived from an oxoacid (organic or inorganic) in which at least one hydroxyl group () is replaced by an alkoxy group (), as in the substitution reaction of a carboxylic acid and an alcohol. Glycerides are fatty acid esters of glycerol; they are important in biology, being one of the main classes of lipids and comprising the bulk of animal fats and vegetable oils. Esters typically have a pleasant smell; those of low molecular weight are commonly used as fragrances and are found in essential oils and pheromones. They perform as high-grade solvents for a broad array of plastics, plasticizers, resins, and lacquers, and are one of the largest classes of synthetic lubricants on the commercial market. Polyesters are important plastics, with monomers linked by ester moieties. Phosphoesters form the backbone of DNA molecules. Nitrate esters, such as nitroglycerin, are known for their explosive properties. '' Nomenclature Etymology ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
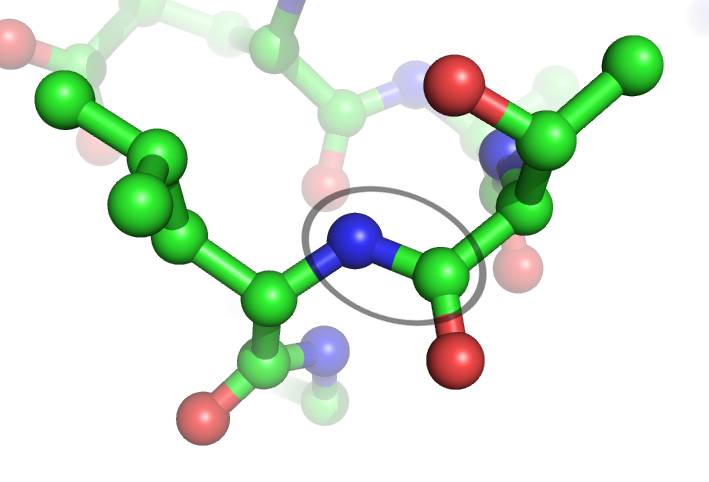
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a ''dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non- side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
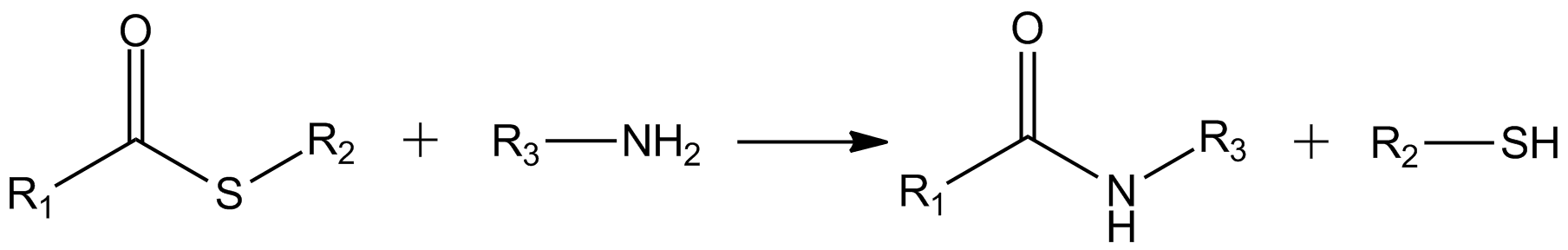
Thioester
In organic chemistry, thioesters are organosulfur compounds with the functional group . They are analogous to carboxylate esters () with the sulfur in the thioester playing the role of the linking oxygen in the carboxylate ester, as implied by the ''thio-'' prefix. They are the product of esterification between a carboxylic acid () and a thiol (). In biochemistry, the best-known thioesters are derivatives of coenzyme A, e.g., acetyl-CoA.Matthys J. Janssen "Carboxylic Acids and Esters" in PATAI's Chemistry of Functional Groups: Carboxylic Acids and Esters, Saul Patai, Ed. John Wiley, 1969, New York: pp. 705–764. Synthesis The most typical route to thioester involves the reaction of an acid chloride with an alkali metal salt of a thiol: :RSNa + R'COCl -> R'COSR + NaCl Another common route entails the displacement of halides by the alkali metal salt of a thiocarboxylic acid. For example, thioacetate esters are commonly prepared by alkylation of potassium thioacetate: :CH3COSK + ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. It is encoded by the codon AUG. Methionine is also an important part of angiogenesis, the growth of new blood vessels. Supplementation may benefit those suffering from copper poisoning. Overconsumption of methionine, the methyl group donor in DNA methylation, is related to cancer growth in a number of studies. Methionine was first isolated in 1921 by John Howard Mueller. Biochemical details Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the protonat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |