|
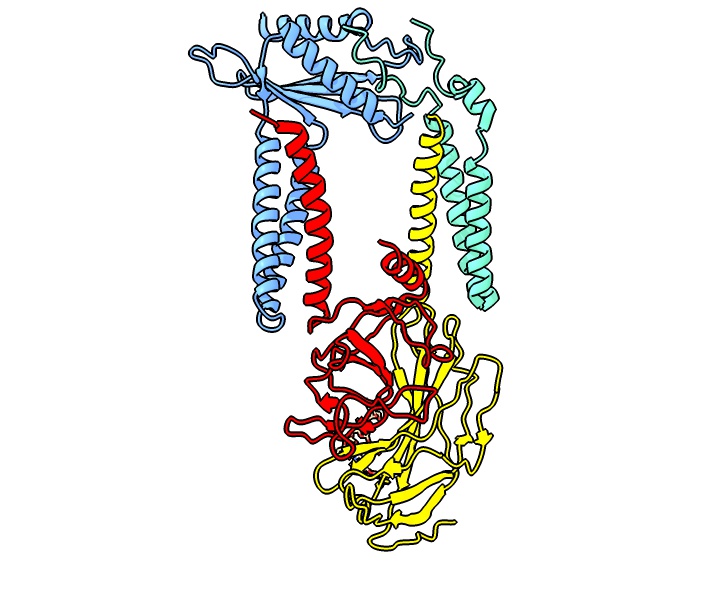
Translocon
The translocon (also known as a translocator or translocation channel) is a complex of proteins associated with the translocation of polypeptides across membranes. In eukaryotes the term translocon most commonly refers to the complex that transports nascent polypeptides with a targeting signal sequence into the interior (cisternal or lumenal) space of the endoplasmic reticulum (ER) from the cytosol. This translocation process requires the protein to cross a hydrophobic lipid bilayer. The same complex is also used to integrate nascent proteins into the membrane itself (membrane proteins). In prokaryotes, a similar protein complex transports polypeptides across the (inner) plasma membrane or integrates membrane proteins. In either case, the protein complex are formed from Sec proteins (Sec: secretory), with the hetrotrimeric Sec61 being the channel. In prokaryotes, the homologous channel complex is known as SecYEG. This article focuses on the cell's native translocons, but pathogens ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligosaccharyl Transferase
Oligosaccharyltransferase or OST () is a membrane protein complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc= ''N''-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X- Ser or Asn-X- Thr where X is any amino acid except proline. This sequence is called a glycosylation ''sequon.'' The reaction catalyzed by OST is the central step in the ''N''-linked glycosylation pathway. Location OST is a component of the translocon in the endoplasmic reticulum (ER) membrane. A lipid-linked core-oligosaccharide is assembled at the membrane of the endoplasmic reticulum and transferred to selected asparagine residues of nascent polypeptide chains by the oligosaccharyl transferase complex. The active site of OST is located about 4 nm from the lumenal face of the ER membrane. It usually acts during translation as the n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. The two types of ER share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of cells contain different ratios of the two types of ER depending on the activities of the cell. RER is found mainly toward the nucleus of cell and SER towards the cell membrane or plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Targeting
:''This article deals with protein targeting in eukaryotes unless specified otherwise.'' Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases. History In 1970, Günter Blobel conducted experiments on protein translocation across membranes. Blobel, then an assistant professor at Rockefeller University, built upon the work of his colleague George Palade. Palade had previously demonstrated that non-secreted proteins were translated by free ribosomes in the cytosol, while secreted proteins (and target proteins, in general) were ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal-recognition Particle
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein (protein- RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane in prokaryotes. History The function of SRP was discovered by the study of processed and unprocessed secretory proteins, particularly immunoglobulin light chains; and bovine preprolactin. Newly synthesized proteins in eukaryotes carry N-terminal hydrophobic Signal peptide, signal sequences, which are bound by SRP when they emerge from the ribosome. Mechanism In eukaryotes, SRP binds to the signal sequence of a newly synthesized peptide as it emerges from the ribosome. This binding leads to the slowing of protein synthesis known as "elongation arrest", a conserved function of SRP that facilitates the coupling of the protein translation and the protein translocation processes. SRP then targets this entire complex (the ribosome-nascent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Recognition Particle
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein ( protein- RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane in prokaryotes. History The function of SRP was discovered by the study of processed and unprocessed secretory proteins, particularly immunoglobulin light chains; and bovine preprolactin. Newly synthesized proteins in eukaryotes carry N-terminal hydrophobic Signal peptide, signal sequences, which are bound by SRP when they emerge from the ribosome. Mechanism In eukaryotes, SRP binds to the signal sequence of a newly synthesized peptide as it emerges from the ribosome. This binding leads to the slowing of protein synthesis known as "elongation arrest", a conserved function of SRP that facilitates the coupling of the protein translation and the protein translocation processes. SRP then targets this entire complex (the ribosome-n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sec61
Sec61, termed SecYEG in prokaryotes, is a membrane protein complex found in all domains of life. As the core component of the translocon, it transports proteins to the endoplasmic reticulum in eukaryotes and out of the cell in prokaryotes. It is a doughnut-shaped pore through the membrane with 3 different subunits (heterotrimeric), SecY (α), SecE (γ), and SecG (β). It has a region called the plug that blocks transport into or out of the ER. This plug is displaced when the hydrophobic region of a nascent polypeptide interacts with another region of Sec61 called the seam, allowing translocation of the polypeptide into the ER lumen. Structure Much of the knowledge on the structure of the SecY/Sec61α pore comes from an X-ray crystallography structure of its archaeal version. The large SecY subunit consists of two halves, trans-membrane segments 1-5 and trans-membrane segments 6-10. They are linked at the extracellular side by a loop between trans-membrane segments 5 and 6. Sec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoplasmic-reticulum-associated Protein Degradation
Endoplasmic-reticulum-associated protein degradation (ERAD) designates a cellular pathway which targets misfolded proteins of the endoplasmic reticulum for ubiquitination and subsequent degradation by a protein-degrading complex, called the proteasome. Mechanism The process of ERAD can be divided into three steps: Recognition of misfolded or mutated proteins in the endoplasmic reticulum The recognition of misfolded or mutated proteins depends on the detection of substructures within proteins such as exposed hydrophobic regions, unpaired cysteine residues and immature glycans. In mammalian cells for example, there exists a mechanism called glycan processing. In this mechanism, the lectin-type chaperones calnexin/calreticulin (CNX/CRT) provide immature glycoproteins the opportunity to reach their native conformation. They can do this by way of reglucosylating these glycoproteins by an enzyme called UDP-glucose-glycoprotein glucosyltransferase also known as UGGT. Terminal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Helix
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SecA
The SecA protein is a cell membrane associated subunit of the eubacterial Sec or Type II secretory pathway, a system which is responsible for the secretion of proteins through the cell membrane. Within this system the SecA ATPase forms a translocase complex with the SecYEG channel, thereby driving the movement of the protein substrate across the membrane. Structure SecA is a complex protein whose structure consists of six characterized domains that can explain SecA's capabilities to bind substrates and to move them. The following five domains seem to be present in all SecA proteins that have been structurally analyzed so far. DEAD motor domain This amino acid domain is subdivided into the two nucleotide binding folds 1 and 2 (NBF1 and NBF2) where ATP is bound and hydrolyzed. The chemical energy from the phosphodiester bonds results in a conformational change which is transferred to other domains (especially the HWD and the PPXD domains) which consequently mechanically mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Immunoglobulin Protein
Binding immunoglobulin protein (BiP) also known as 78 kDa glucose-regulated protein (GRP-78) or heat shock 70 kDa protein 5 (HSPA5) is a protein that in humans is encoded by the ''HSPA5'' gene. BiP is a HSP70 molecular chaperone located in the lumen of the endoplasmic reticulum (ER) that binds newly synthesized proteins as they are translocated into the ER, and maintains them in a state competent for subsequent folding and oligomerization. BiP is also an essential component of the translocation machinery and plays a role in retrograde transport across the ER membrane of aberrant proteins destined for degradation by the proteasome. BiP is an abundant protein under all growth conditions, but its synthesis is markedly induced under conditions that lead to the accumulation of unfolded polypeptides in the ER. Structure BiP contains two functional domains: a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD). The NBD binds and hydrolyzes ATP, and the SBD binds ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SRP Receptor
Signal recognition particle (SRP) receptor, also called the docking protein, is a dimer composed of 2 different subunits that are associated exclusively with the rough ER in mammalian cells. Its main function is to identify the SRP units. SRP (signal recognition particle) is a molecule that helps the ribosome-mRNA-polypeptide complexes to settle down on the membrane of the endoplasmic reticulum. The eukaryotic SRP receptor (termed SR) is a heterodimer of SR-alpha (70 kDa; SRPRA) and SR-beta (25 kDa; SRPRB), both of which contain a GTP-binding domain, while the prokaryotic SRP receptor comprises only the monomeric loosely membrane-associated SR-alpha homologue FtsY (). SRX domain SR-alpha regulates the targeting of SRP-ribosome-nascent polypeptide complexes to the translocon. SR-alpha binds to the SRP54 subunit of the SRP complex. The SR-beta subunit is a transmembrane GTPase that anchors the SR-alpha subunit (a peripheral membrane GTPase) to the ER membrane. SR-beta interac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptidase
Signal peptidases are enzymes that convert secretory and some membrane proteins to their mature or pro forms by cleaving their signal peptides from their N-termini. Signal peptidases were initially observed in endoplasmic reticulum (ER)-derived membrane fractions isolated from mouse myeloma cells. The key observation by César Milstein and colleagues was that immunoglobulin light chains were produced in a higher molecular weight form, which became processed by the ER membrane fraction. This finding was directly followed by the discovery of the translocation machinery. Signal peptidases are also found in prokaryotes as well as the protein import machinery of mitochondria and chloroplasts. All signal peptidases described so far are serine proteases. The active site that endoproteolytically cleaves signal peptides from translocated precursor proteins is located at the extracytoplasmic site of the membrane. The eukaryotic signal peptidase is an integral membrane protein Me ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.png)