|
Regulator Gene
A regulator gene, regulator, or regulatory gene is a gene involved in controlling the expression of one or more other genes. Regulatory sequences, which encode regulatory genes, are often at the five prime end (5') to the start site of transcription of the gene they regulate. In addition, these sequences can also be found at the three prime end (3') to the transcription start site. In both cases, whether the regulatory sequence occurs before (5') or after (3') the gene it regulates, the sequence is often many kilobases away from the transcription start site. A regulator gene may encode a protein, or it may work at the level of RNA, as in the case of genes encoding microRNAs. An example of a regulator gene is a gene that codes for a repressor protein that inhibits the activity of an operator (a gene which binds repressor proteins thus inhibiting the translation of RNA to protein via RNA polymerase). In prokaryotes, regulator genes often code for repressor proteins. Repress ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Regulatory Network
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo). The regulator can be DNA, RNA, protein or any combination of two or more of these three that form a complex, such as a specific sequence of DNA and a transcription factor to activate that sequence. The interaction can be direct or indirect (through transcribed RNA or translated protein). In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enzym ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Module
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Silencer (DNA)
In genetics, a silencer is a DNA sequence capable of binding transcription regulation factors, called repressors. DNA contains genes and provides the template to produce messenger RNA (mRNA). That mRNA is then translated into proteins. When a repressor protein binds to the silencer region of DNA, RNA polymerase is prevented from transcribing the DNA sequence into RNA. With transcription blocked, the translation of RNA into proteins is impossible. Thus, silencers prevent genes from being expressed as proteins. RNA polymerase, a DNA-dependent enzyme, transcribes the DNA sequences, called nucleotides, in the 3' to 5' direction while the complementary RNA is synthesized in the 5' to 3' direction. RNA is similar to DNA, except that RNA contains uracil, instead of thymine, which forms a base pair with adenine. An important region for the activity of gene repression and expression found in RNA is the 3' untranslated region. This is a region on the 3' terminus of RNA that will not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. Al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
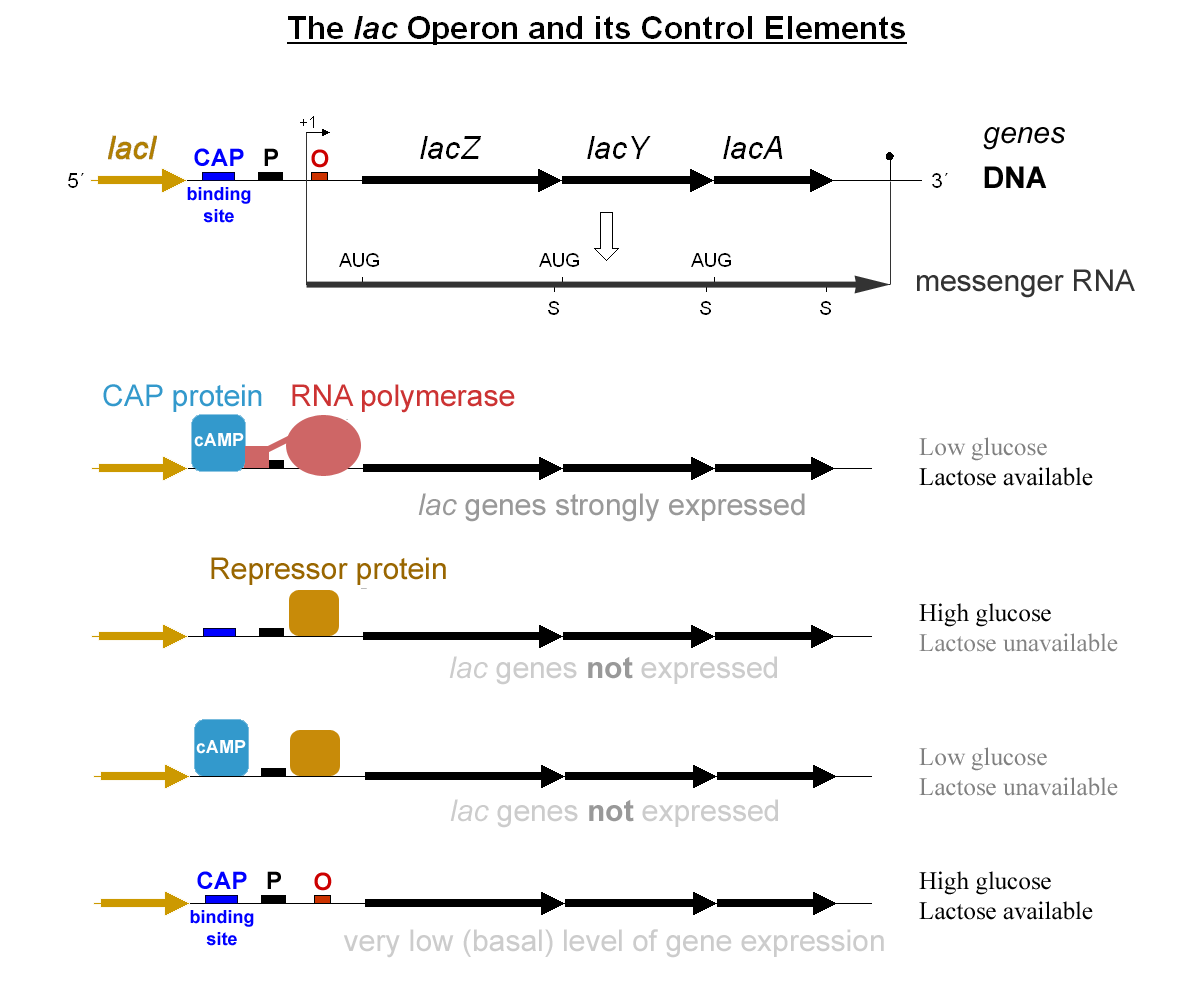
Lac Operon
The ''lactose'' operon (''lac'' operon) is an operon required for the transport and metabolism of lactose in ''E. coli'' and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the ''lac'' operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the ''lac'' operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the ''lac'' operon won them the Nobel Prize in Physiology in 1965. Bacterial operons are polycistronic transcripts that are able to produce multiple proteins from one mRNA transcript. In this case, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
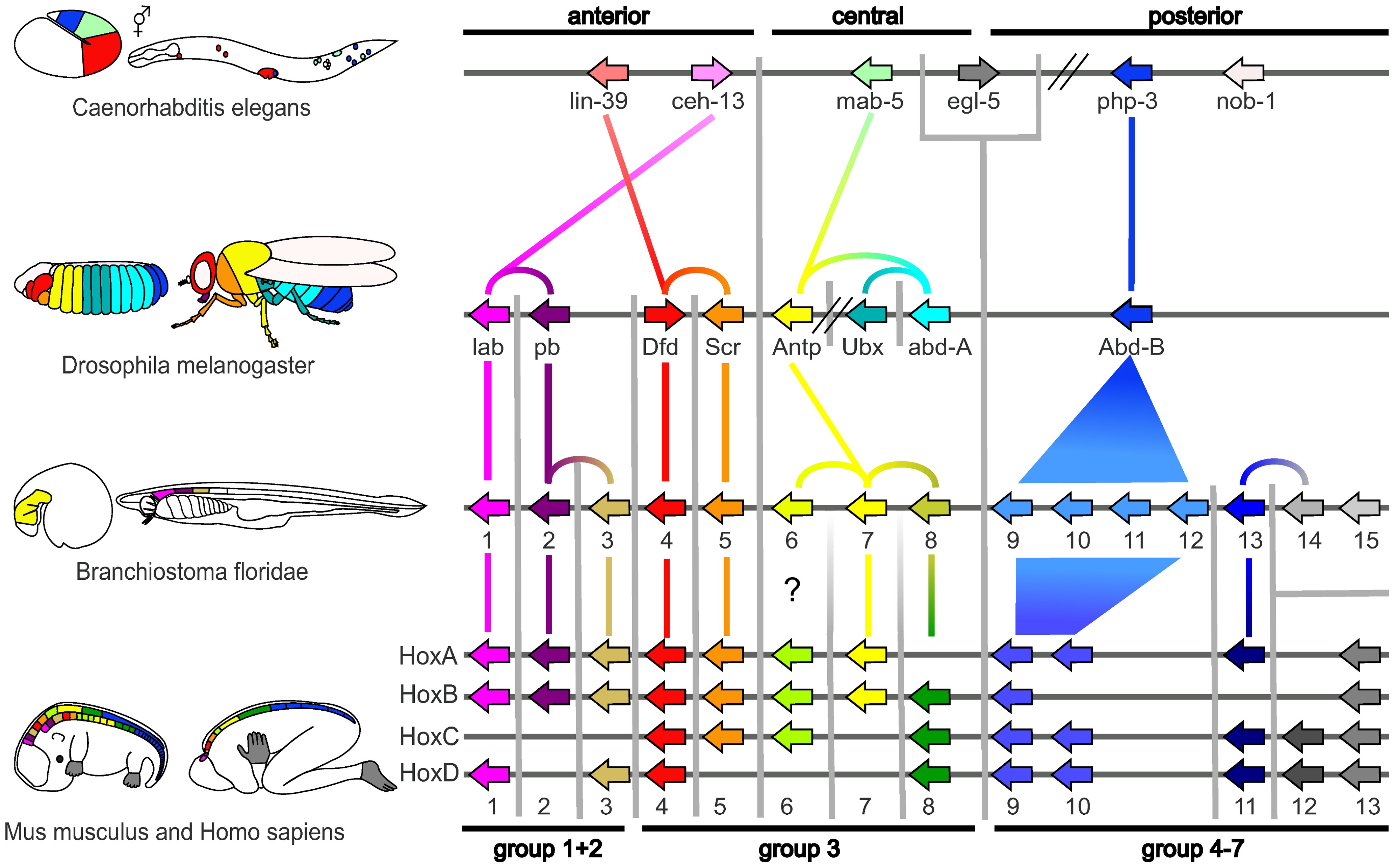
Evolutionary Developmental Biology
Evolutionary developmental biology (informally, evo-devo) is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved. The field grew from 19th-century beginnings, where embryology faced a mystery: zoologists did not know how embryonic development was controlled at the molecular level. Charles Darwin noted that having similar embryos implied common ancestry, but little progress was made until the 1970s. Then, recombinant DNA technology at last brought embryology together with molecular genetics. A key early discovery was of homeotic genes that regulate development in a wide range of eukaryotes. The field is composed of multiple core evolutionary concepts. One is deep homology, the finding that dissimilar organs such as the eyes of insects, vertebrates and cephalopod molluscs, long thought to have evolved separately, are controlled by similar genes such as ''pax-6'', from the evo-devo gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Gene Expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color formation of maize see ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lac Operon
The ''lactose'' operon (''lac'' operon) is an operon required for the transport and metabolism of lactose in ''E. coli'' and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the ''lac'' operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the ''lac'' operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the ''lac'' operon won them the Nobel Prize in Physiology in 1965. Bacterial operons are polycistronic transcripts that are able to produce multiple proteins from one mRNA transcript. In this case, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catabolite Activator Protein
Catabolite activator protein (CAP; also known as cAMP receptor protein, CRP) is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus (CAPN, residues 1–138) and a DNA-binding domain at the C-terminus (DBD, residues 139–209). Two cAMP (cyclic AMP) molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open conformation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively cata ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activator Protein
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |