|
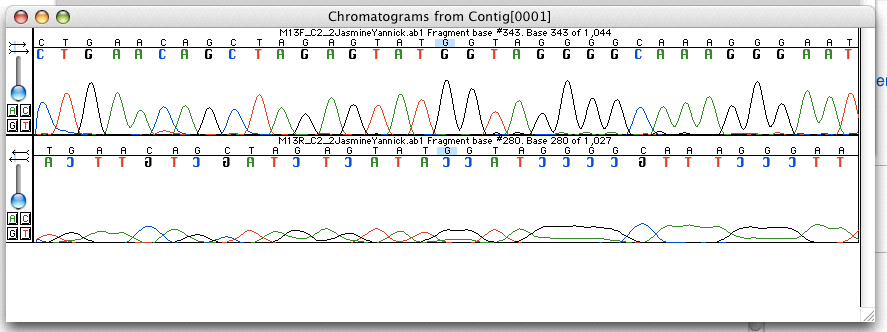
Pyrosequencing
Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase. Pyrosequencing relies on light detection based on a chain reaction when pyrophosphate is released. Hence, the name pyrosequencing. The principle of pyrosequencing was first described in 1993 by, Bertil Pettersson, Mathias Uhlén, Mathias Uhlen and Pål Nyrén, Pål Nyren by combining the solid phase sequencing method using streptavidin coated magnetic beads with recombinant DNA polymerase lacking 3´to 5´exonuclease activity (proof-reading) and luminescence detection using the Luciferase, firefly luciferase enzyme. A mixture of three enzymes (DNA polymerase, Sulfate adenylyltransferase, ATP sulfurylase and firefly luciferase) and a nucleotide (Nucleoside triphosphate, dNTP) are added to single stranded DNA to be sequenced and the incorporation o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, Genographic Project, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid advancements in DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
454 Life Sciences
454 Life Sciences was a biotechnology company based in Branford, Connecticut that specialized in high-throughput DNA sequencing. It was acquired by Roche in 2007 and shut down by Roche in 2013 when its technology became noncompetitive, although production continued until mid-2016. History 454 Life Sciences was founded by Jonathan Rothberg and was originally known as 454 Corporation, a subsidiary of CuraGen. For their method for low-cost gene sequencing, 454 Life Sciences was awarded the Wall Street Journal's Gold Medal for Innovation in the Biotech-Medical category in 2005. The name 454 was the code name by which the project was referred to at CuraGen, and the numbers have no known special meaning. In November 2006, Rothberg, Michael Egholm, and colleagues at 454 published a cover article with Svante Pääbo in Nature describing the first million base pairs of the Neanderthal genome, and initiated the Neanderthal Genome Project to complete the sequence of the Neanderthal ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Assembly
In bioinformatics, sequence assembly refers to aligning and merging fragments from a longer DNA sequence in order to reconstruct the original sequence. This is needed as DNA sequencing technology might not be able to 'read' whole genomes in one go, but rather reads small pieces of between 20 and 30,000 bases, depending on the technology used. Typically, the short fragments (reads) result from shotgun sequencing genomic DNA, or gene transcript ( ESTs). The problem of sequence assembly can be compared to taking many copies of a book, passing each of them through a shredder with a different cutter, and piecing the text of the book back together just by looking at the shredded pieces. Besides the obvious difficulty of this task, there are some extra practical issues: the original may have many repeated paragraphs, and some shreds may be modified during shredding to have typos. Excerpts from another book may also be added in, and some shreds may be completely unrecognizable. Typ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomics
Genomics is an interdisciplinary field of molecular biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of ''individual'' genes and their roles in inheritance, genomics aims at the collective characterization and quantification of ''all'' of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemiluminescence
Chemiluminescence (also chemoluminescence) is the emission of light (luminescence) as the result of a chemical reaction, i.e. a chemical reaction results in a flash or glow of light. A standard example of chemiluminescence in the laboratory setting is the luminol test. Here, blood is indicated by luminescence upon contact with iron in hemoglobin. When chemiluminescence takes place in living organisms, the phenomenon is called bioluminescence. A light stick emits light by chemiluminescence. Physical description As in many chemical reactions, chemiluminescence starts with the combining of two compounds, say A and B, to give a product C. Unlike most chemical reactions, the product C converts to a further product, which is produced in an electronically excited state often indicated with an asterisk: : A + B → C : C → D* D* then emits a photon (''h''ν), to give the ground state of D: I : D* → D + ''h''ν In theory, one photon of light should be given off for each mol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apyrase
Apyrase (, ''ATP-diphosphatase'', adenosine diphosphatase, ''ADPase'', ''ATP diphosphohydrolase'') is a calcium-activated plasma membrane-bound enzyme (magnesium can also activate it) () that catalyses the hydrolysis of ATP to yield AMP and inorganic phosphate. Two isoenzymes are found in commercial preparations from ''S. tuberosum''. One with a higher ratio of substrate selectivity for ATP:ADP (approx 10) and another with no selectivity (ratio 1). It can also act on ADP and other nucleoside triphosphates and diphosphates with the general reaction being NTP -> NDP + Pi -> NMP + 2Pi. This is the same activity that has been employed in the degradation of unincorporated nucleosides during pyrosequencing. The salivary apyrases of blood-feeding arthropods are nucleotide hydrolysing enzymes that are implicated in the inhibition of host platelet aggregation through the hydrolysis of extracellular adenosine diphosphate Adenosine diphosphate (ADP), also known as adenosine pyrophosp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mostafa Ronaghi
Mostafa Ronaghi (; born 1968) is an Iranian molecular biologist, specializing in DNA sequencing methodology. He earned his Ph.D. from the Royal Institute of Technology in Sweden in 1998. , he is the Chief Technology Officer and Senior Vice President at Illumina. Prior to this position, he was a principal investigator and Senior Research Associate at the Stanford Genome Technology Center at Stanford University, focusing on developing analytical techniques for molecular diagnostics. He is principal investigator for several grants including grants from the National Human Genome Research Institute (NHGRI), part of the National Institutes of Health for the development of array-based Pyrosequencing. In 1998, he described together with Pål Nyren anMathias Uhlena solution-based variant of the pyrosequencing technology and co-founded Pyrosequencing AB (renamed to Biotage in 2003). He co-invented Molecular Inversion Probe assay and co-founded ParAllele BioScience to develop this multip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATP Sulfurylase
In enzymology, a sulfate adenylyltransferase () is an enzyme that catalyzes the chemical reaction : ATP + sulfate ⇌ pyrophosphate + adenylyl sulfate Thus, the two substrates of this enzyme are ATP and sulfate, whereas its two products are pyrophosphate and adenylyl sulfate. This enzyme belongs to the family of transferases, specifically those transferring phosphorus-containing nucleotide groups ( nucleotidyltransferases). The systematic name of this enzyme class is ATP:sulfate adenylyltransferase. Other names in common use include adenosine-5'-triphosphate sulfurylase, adenosinetriphosphate sulfurylase, adenylylsulfate pyrophosphorylase, ATP sulfurylase, ATP-sulfurylase, and sulfurylase. This enzyme participates in 3 metabolic pathways: purine metabolism, selenoamino acid metabolism, and sulfur metabolism. Some sulfate adenylyltransferases are part of a bifunctional polypeptide chain associated with adenosyl phosphosulfate (APS) kinase. Both enzymes are required for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pål Nyrén
Pål Nyrén (born 1955) is a biochemistry professor at the Royal Institute of Technology (KTH), Stockholm. He is most famous for developing the pyrosequencing method for DNA sequencing. Career *1999 Professor in Biochemistry, KTH, Stockholm *1997 Founder of the company Biotage AB (former Pyrosequencing AB) *1988 Associate professor (Docent) Biochemistry, Stockholm University *1985-86 Postdoc at LMB, MRC, Cambridge, G.B. with prof John Walker *1985 PhD (Tekn. Doktor) Biochemistry, Stockholm University (Thesis title: "The proton pumping pyrophosphatase from ''Rhodospirillum rubrum''") *1981 MSc (Civ. ing.) Chemical Engineering, KTH, Stockholm Recognition *2013 winner of the European Inventor Award in the SMEs category |
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondrial DNA, mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of DNA sequencing, gene sequencing at Single-nucleotide polymorphism, SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. Whole genome sequencing should not be confused with DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
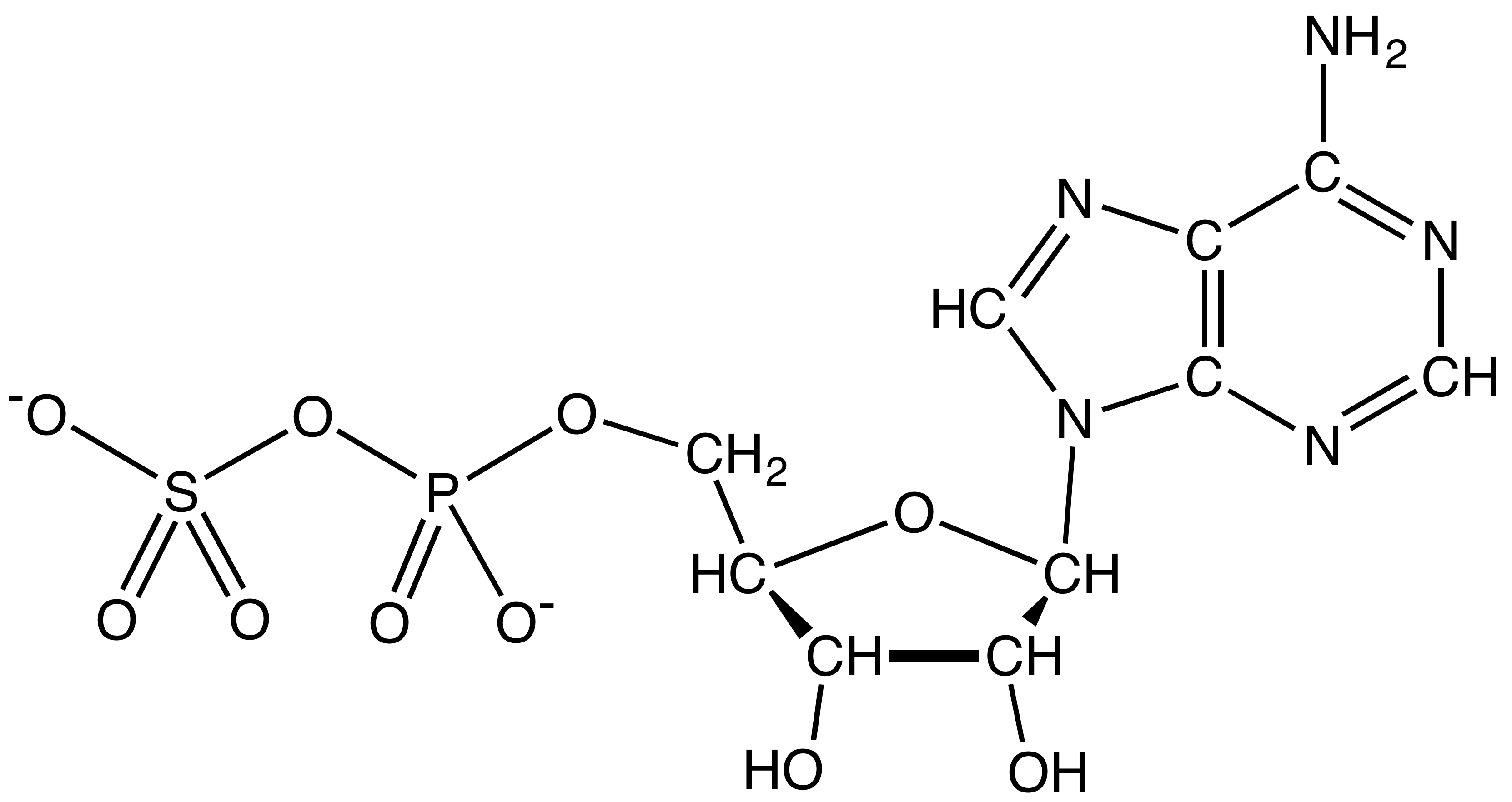
Adenosine 5´ Phosphosulfate
3′-Phosphoadenosine-5′-phosphosulfate (PAPS) is a derivative of adenosine monophosphate (AMP) that is phosphorylated at the 3′ position and has a sulfate group attached to the 5′ phosphate. It is the most common coenzyme in sulfotransferase reactions and hence part of sulfation pathways. It is endogenously synthesized by organisms via the phosphorylation of adenosine 5′-phosphosulfate (APS), an intermediary metabolite. In humans such reaction is performed by bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthases (PAPSS1 and PAPSS2) using Adenosine triphosphate, ATP as the phosphate donor. Formation and reduction APS and PAPS are intermediates in the reduction of sulfate to sulfite, an exothermic conversion that is carried out by sulfate-reducing bacteria. In these organisms, sulfate serves as an electron acceptor, akin to the use of O2 as an electron acceptor by aerobic organisms. Sulfate is not reduced directly but must be activated by the formation of APS or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |