|
Promoter Activity
Promoter activity is a term that encompasses several meanings around the process of gene expression from regulatory sequences —promoter (genetics), promoters and Enhancer (genetics), enhancers. Gene expression has been commonly characterized as a measure of how much, how fast, when and where this process happens. Promoters and enhancers are required for controlling where and when a specific gene is transcribed. Traditionally the measure of gene products (i.e. mRNA, proteins, etc.) has been the major approach of measure promoter activity. However, this method confront with two issues: the stochastic nature of the gene expression and the lack of mechanistic interpretation of the thermodynamical process involved in the promoter activation. The actual developments in metabolomics product of developments of next-generation sequencing technologies and molecular structural analysis have enabled the development of more accurate models of the process of promoter activation (e.g. the sig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics ( phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutation is the ultima ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization ( body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-Regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ty ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glossary Of Gene Expression Terms
A glossary (from grc, γλῶσσα, ''glossa''; language, speech, wording) also known as a vocabulary or clavis, is an alphabetical list of terms in a particular domain of knowledge with the definitions for those terms. Traditionally, a glossary appears at the end of a book and includes terms within that book that are either newly introduced, uncommon, or specialized. While glossaries are most commonly associated with non-fiction books, in some cases, fiction novels may come with a glossary for unfamiliar terms. A bilingual glossary is a list of terms in one language defined in a second language or glossed by synonyms (or at least near-synonyms) in another language. In a general sense, a glossary contains explanations of concepts relevant to a certain field of study or action. In this sense, the term is related to the notion of ontology. Automatic methods have been also provided that transform a glossary into an ontology or a computational lexicon. History In medieval Eu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish '' Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reporter Gene
In molecular biology, a reporter gene (often simply reporter) is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population. Common reporter genes To introduce a reporter gene into an organism, scientists place the reporter gene and the gene of interest in the same DNA construct to be inserted into the cell or organism. For bacteria or prokaryotic cells in culture, this is usually in the form of a circular DNA molecule called a plasmid. For viruses, this is known as a viral vector. It is important to use a reporter gene that is not natively expressed in the cell or organism under study, since ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
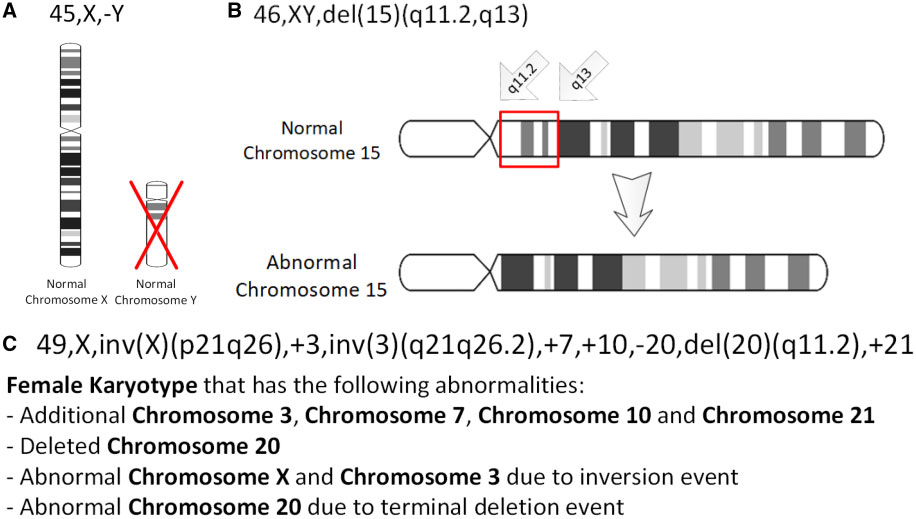
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Footprinting
Phylogenetic footprinting is a technique used to identify transcription factor binding sites (TFBS) within a non-coding region of DNA of interest by comparing it to the orthologous sequence in different species. When this technique is used with a large number of closely related species, this is called phylogenetic shadowing. Researchers have found that non-coding pieces of DNA contain binding sites for regulatory proteins that govern the spatiotemporal expression of genes. These transcription factor binding sites (TFBS), or regulatory motifs, have proven hard to identify, primarily because they are short in length, and can show sequence variation. The importance of understanding transcriptional regulation to many fields of biology has led researchers to develop strategies for predicting the presence of TFBS, many of which have led to publicly available databases. One such technique is Phylogenetic Footprinting. Phylogenetic footprinting relies upon two major concepts: # The fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life— eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sigma Factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP). The sigma factor, together with RNA polymerase, is known as the RNA polymerase holoenzyme. Every molecule of RNA polymerase holoenzyme contains exactly one sigma factor subunit, which in the model bacterium ''Escherichia coli'' is one of those listed below. The number of sigma f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |