|
Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Histone Methyltransferase
Histone methyltransferases (HMT) are histone-modifying enzymes (e.g., histone-lysine N-methyltransferases and histone-arginine N-methyltransferases), that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific (which can be SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain containing or non-SET domain containing) and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group. The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
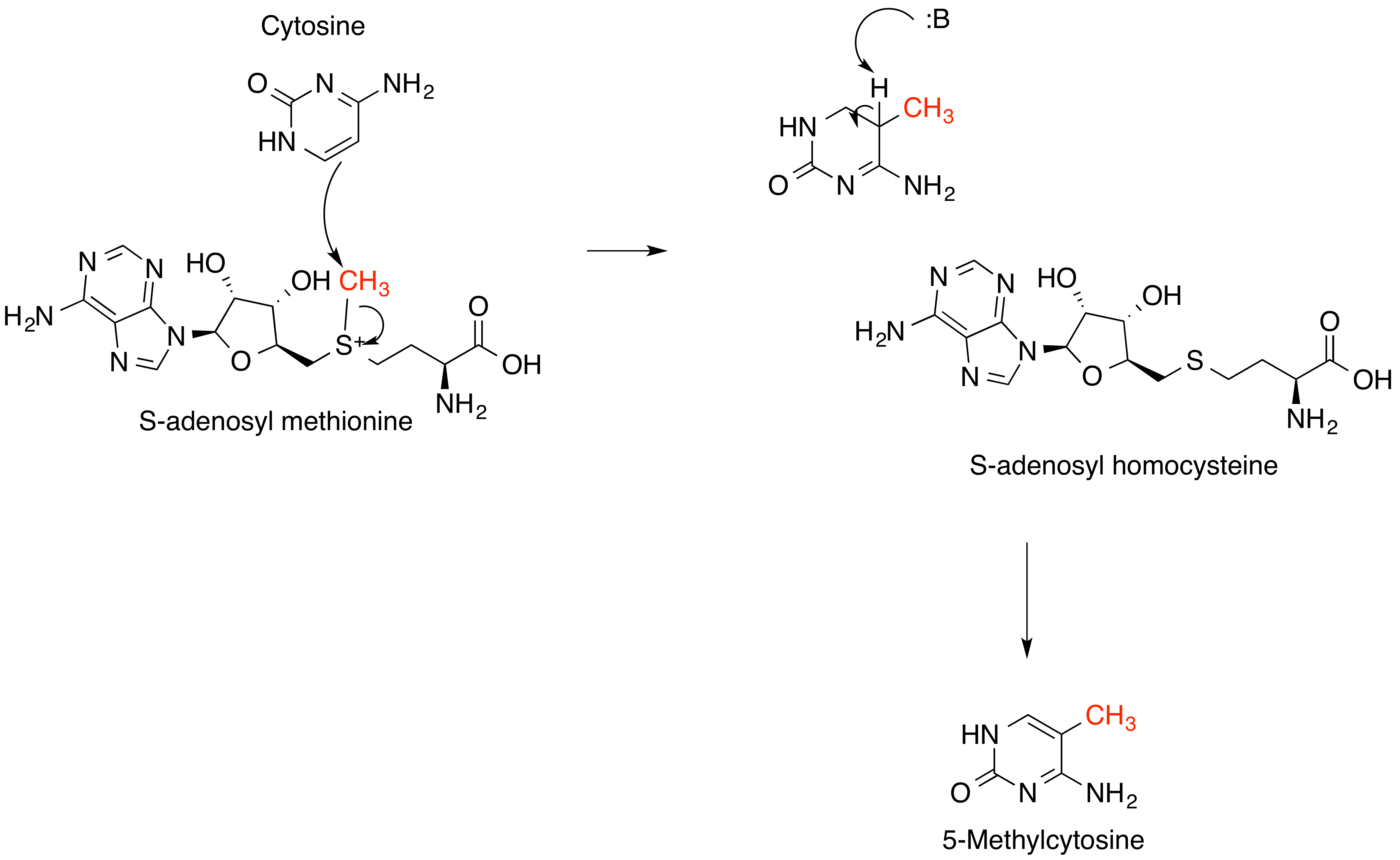
Methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen#Compounds, hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and biology. In biological systems, methylation is Catalysis, catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of Protein#Functions, protein function, and RNA processing. ''In vitro'' methylation of tissue samples is also a way to reduce some histology#Histological Artifacts, histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals and can regulate gene expression, RNA processing, and protein function. It is a key pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
SET7 9
Histone-lysine N-methyltransferase SETD7 is an enzyme that in humans is encoded by the ''SETD7'' gene In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei .... References Further reading * * * * * * * * * * * * {{gene-4-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. Methionine is also involved in angiogenesis and various processes related to DNA transcription, epigenetic expression, and gene regulation. Methionine was first isolated in 1921 by John Howard Mueller. It is Genetic code, encoded by the codon AUG. It was named by Satoru Odake in 1925, as an abbreviation of its structural description 2-amino-4-(methylthio)butanoic acid. Biochemical details Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the proton ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many RNA viruses is composed of negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process is catalyzed by a viral RNA dependent RNA polymerase. Background A DNA transcription unit encoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
SETD6
SET domain containing 6 is a protein in humans that is encoded by the SETD6 gene In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei .... SETD6 monomethylates the RelA subunit of nuclear factor kappa B (NF-κB). RelA mono-methylation at lysine 310 (RelAK310me1) leads to the constitutive repression of RelA target genes by recruiting the PKMT G9a-like protein (GLP), which catalyzes H3K9me2 and leads to chromatin silencing and gene repression. In response to stimulation with TNFa and lipopolysaccharide, phosphorylation of RelA at serine 311 (RelAS311ph) by PKCzeta physically blocks the interaction between GLP and RelAK310me1, leading to transcription activation. PAK4 Methylation by SETD6 Promotes the Activation of the Wnt/β-Catenin Pathway. SETD6 binds and methylates PAK4 both in vitro a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA repair#DNA damage, DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30 nm fiber. ''Current opinion in cell biolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Genomic Imprinting
Genomic imprinting is an epigenetic phenomenon that causes genes to be expressed or not, depending on whether they are inherited from the female or male parent. Genes can also be partially imprinted. Partial imprinting occurs when alleles from both parents are differently expressed rather than complete expression and complete suppression of one parent's allele. Forms of genomic imprinting have been demonstrated in fungi, plants and animals. In 2014, there were about 150 imprinted genes known in mice and about half that in humans. As of 2019, 260 imprinted genes have been reported in mice and 228 in humans. Genomic imprinting is an inheritance process independent of the classical Mendelian inheritance. It is an epigenetic process that involves DNA methylation and histone methylation without altering the genetic sequence. These epigenetic marks are established ("imprinted") in the germline (sperm or egg cells) of the parents and are maintained through mitotic cell divisions in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Regulation Of Gene Expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from Transcriptional regulation, transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
S-Adenosyl Methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952. In bacteria, SAM is bound by the SAM riboswitch, which regulates genes involved in methionine or cysteine biosynthesis. In eukaryotic cells, SAM serves as a regulator of a variety of processes including DNA, tRNA, and rRNA methylation; immune response; amino acid metabolism; transsulfuration; and more. In plants, SAM is crucial to the biosynthesis of ethylene, an important plant hormone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |