|
Haplogroup BT
Haplogroup BT M91, also known as Haplogroup A1b2 (and formerly as A4, BR and BCDEF), is a Y-chromosome haplogroup. BT is a subclade of haplogroup A1b (P108) and a sibling of the haplogroup A1b1 (L419/PF712). Distribution Basal BT* has not been documented in any living individuals or ancient remains. Later Stone Age individuals excavated at Fingira Rock, Malawi, dated to around 6100 years ago (2/2 males), and at Mount Hora, Malawi, dated to around 8000 years ago (1/1 males), all belonged to Y haplogroup BT(xCT) (i.e. they did not belong to haplogroup CT but may have belonged to haplogroup B). No definite examples of BT(xCF,DE) – i.e. members of BT outside the only two known branches of CT, namely haplogroups CF and DE – have been identified. In some cases, because testing is undertaken only for geographically and historically likely haplogroups, the data required to identify a precise subclade has not been collected and/or recorded. For instance, research published in 201 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup A (Y-DNA)
Haplogroup A is a human Y-chromosome DNA haplogroup, which includes all living human Y chromosomes. Bearers of extant sub-clades of haplogroup A are almost exclusively found in Africa (or among descendants of populations which have recently left Africa), in contrast with haplogroup BT, bearers of which participated in the Out of Africa migration of anatomically modern humans. The known branches of haplogroup A are A00, A0, A1a, and A1b1; these branches are only very distantly related, and are not more closely related to each other than they are to haplogroup BT. Origin There are terminological difficulties, but as "haplogroup A" has come to mean "the foundational haplogroup" (viz. of contemporary human population), haplogroup A is not defined by any mutation but refers to any haplogroup which is not descended from the haplogroup BT, i.e. defined by the absence of the defining mutation of that group (M91). By this definition, haplogroup A includes all mutations that took pla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
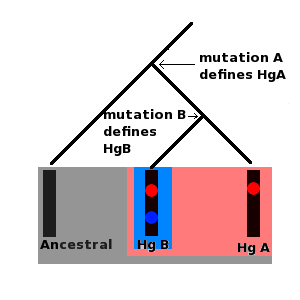
Haplogroup A Tree
A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup ( haploid from the el, ἁπλοῦς, ''haploûs'', "onefold, simple" and en, group) is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism mutation. More specifically, a haplogroup is a combination of alleles at different chromosomal regions that are closely linked and that tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual. Each haplogroup originates from, and remains part of, a preceding single haplogroup (or paragroup). As such, any related group of haplogroups may be precisely modelled as a nested hierarchy, in which each set ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Y-DNA Haplogroups By Ethnic Group
The various ethnolinguistic groups found in the Caucasus, Central Asia, Europe, the Middle East, North Africa and/or South Asia demonstrate differing rates of particular Y-DNA haplogroups. In the table below, the first two columns identify ethnolinguistic groups. Subsequent columns represent the sample size (''n'') of the study or studies cited, and the percentage of each haplogroup found in that particular sample. (Data from studies conducted before 2004 may be inaccurate or a broad estimate, due to obsolete haplogroup naming systems – e.g. the former ''Haplogroup 2'' included members of the relatively unrelated haplogroups known later as Haplogroup G and macrohaplogroup IJ hich comprises haplogroups I and J) See also * Genetics ** Human mitochondrial DNA haplogroups ** Human genome ** Genetic genealogy ** Genealogical DNA testing ** Race and genetics * Genetic history * Timeline of human evolution ** Recent African origin of modern humans ** Genetic history of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Y-chromosome Haplogroups In Populations Of The World
The following articles are lists of human Y-chromosome DNA haplogroups found in populations around the world. * Y-DNA haplogroups by ethnic group *Y-DNA haplogroups in populations of Europe * Y-DNA haplogroups in populations of the Near East *Y-DNA haplogroups in populations of North Africa * Y-DNA haplogroups in populations of Sub-Saharan Africa * Y-DNA haplogroups in populations of the Caucasus *Y-DNA haplogroups in populations of South Asia *Y-DNA haplogroups in populations of East and Southeast Asia * Y-DNA haplogroups in populations of Central and North Asia *Y-DNA haplogroups in populations of Oceania * Y-DNA haplogroups in indigenous peoples of the Americas * List of haplogroups of historic people See also *Recent African origin of modern humans *Genetic history of the Middle East *Genetics and archaeogenetics of South Asia * Genetic history of Europe *Genetic history of Italy *Genetic history of North Africa * Genetic history of Indigenous peoples of the Americas *Genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subclade
In genetics, a subclade is a subgroup of a haplogroup. Naming convention Although human mitochondrial DNA (mtDNA) and Y chromosome DNA (Y-DNA) haplogroups and subclades are named in a similar manner, their names belong to completely separate systems. mtDNA mtDNA haplogroups are defined by the presence of a series of single-nucleotide polymorphism (SNP) markers in the hypervariable regions and the coding region of mitochondrial DNA. They are named with the capital letters A through Z, with further subclades named using numbers and lower case letters. Y-DNA Y-DNA haplogroups are defined by the presence of a series of SNP markers on the Y chromosome. Subclades are defined by a ''terminal SNP'', the SNP furthest down in the Y chromosome phylogenetic tree. Human Y-DNA The Y Chromosome Consortium (YCC) developed a system of naming major human Y-DNA haplogroups with the capital letters A through T, with further subclades named using numbers and lower case letters (YCC longhand nom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paragroup
Paragroup is a term used in population genetics to describe lineages within a haplogroup that are not defined by any additional unique markers. In human Y-chromosome DNA haplogroups, paragroups are typically represented by an asterisk (*) placed after the main haplogroup. The term "paragroup" is a portmanteau of the terms paraphyletic haplogroup A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup (haploid from the el, ἁπλοῦς, ''haploûs'', "onefold, simple" and en, group) is a group of similar haplotypes that share ... indicating that paragroups form paraphyletic subclades. Apart from the mutations that define the parent haplogroup, paragroups may not possess any additional unique markers. Alternatively paragroups may possess unique markers that have not been discovered. If a unique marker is discovered within a paragroup, the specific lineage is given a unique name and is moved out of the paragroup to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Phylogenetics
Molecular phylogenetics () is the branch of phylogeny that analyzes genetic, hereditary molecular differences, predominantly in DNA sequences, to gain information on an organism's evolutionary relationships. From these analyses, it is possible to determine the processes by which diversity among species has been achieved. The result of a molecular phylogenetic analysis is expressed in a phylogenetic tree. Molecular phylogenetics is one aspect of molecular systematics, a broader term that also includes the use of molecular data in taxonomy and biogeography. Molecular phylogenetics and molecular evolution correlate. Molecular evolution is the process of selective changes (mutations) at a molecular level (genes, proteins, etc.) throughout various branches in the tree of life (evolution). Molecular phylogenetics makes inferences of the evolutionary relationships that arise due to molecular evolution and results in the construction of a phylogenetic tree. History The theoretical f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotype
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup
A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup (haploid from the el, ἁπλοῦς, ''haploûs'', "onefold, simple" and en, group) is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism mutation. More specifically, a haplogroup is a combination of alleles at different chromosomal regions that are closely linked and that tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual. Each haplogroup originates from, and remains part of, a preceding single haplogroup (or paragroup). As such, any related group of haplogroups may be precisely modelled as a nested hierarchy, in which each set (ha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup DE (Y-DNA)
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183. DE is unique because it is distributed in several geographically distinct clusters. An immediate subclade, haplogroup D (also known as D-CTS3946), is mainly found in East Asia, parts of Central Asia, and the Andaman Islands, but also sporadically in West Africa and West Asia. The other immediate subclade, haplogroup E, is common in Africa, and to a lesser extent the Middle East and southern Europe. The most well-known unique event polymorphism (UEP) that defines DE is the Y-chromosome Alu Polymorphism "YAP''"''. The mutation was caused when a strand of DNA, known as Alu, inserted a copy of itself into the Y chromosome. Hence, all Y chromosomes belonging to DE, D, E and their subclades are YAP-positive (YAP+). All Y chromosomes that belong to other haplogroups and subclades are YAP-negative ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplogroup D (Y-DNA)
Haplogroup D or D-CTS3946 is a Y-chromosome haplogroup. Like its relative distant sibling, haplogroup E-M96, D-CTS3946 has the YAP+ unique-event polymorphism, which defines their parent, haplogroup DE. Subclades of haplogroup D-CTS3946 are found primarily in East Asia, though they are also found regularly with low frequency in Central Asia and Southeast Asia, and have also been found sporadically in Western Africa and Western Asia. Overview Haplogroup D was formerly the name of the D lineage D-M174. Varying proposals exist regarding the origin of haplogroup DE, the parent of D, with some suggesting an African and others an Asian origin. But D-M174 was, and generally is, assumed to be of Asian origin and is exclusively found in Asia. However, a study by Haber et al. (2019) identified a haplogroup, termed "D0", in three Nigerians. Defined by the SNP A5580.2, "D0" haplogroup is outside M174, but belongs to the D lineage, shares 7 SNPs with it D-M174 that E lacks, and was dete ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Genealogy
Genetic genealogy is the use of genealogical DNA tests, i.e., DNA profiling and DNA testing, in combination with traditional genealogical methods, to infer genetic relationships between individuals. This application of genetics came to be used by family historians in the 21st century, as DNA tests became affordable. The tests have been promoted by amateur groups, such as surname study groups or regional genealogical groups, as well as research projects such as the Genographic Project. As of 2019, about 30 million people had been tested. As the field developed, the aims of practitioners broadened, with many seeking knowledge of their ancestry beyond the recent centuries, for which traditional pedigrees can be constructed. History The investigation of surnames in genetics can be said to go back to George Darwin, a son of Charles Darwin and Charles' first cousin Emma Darwin. In 1875, George Darwin used surnames to estimate the frequency of first-cousin marriages and calcul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |





.png)
