|
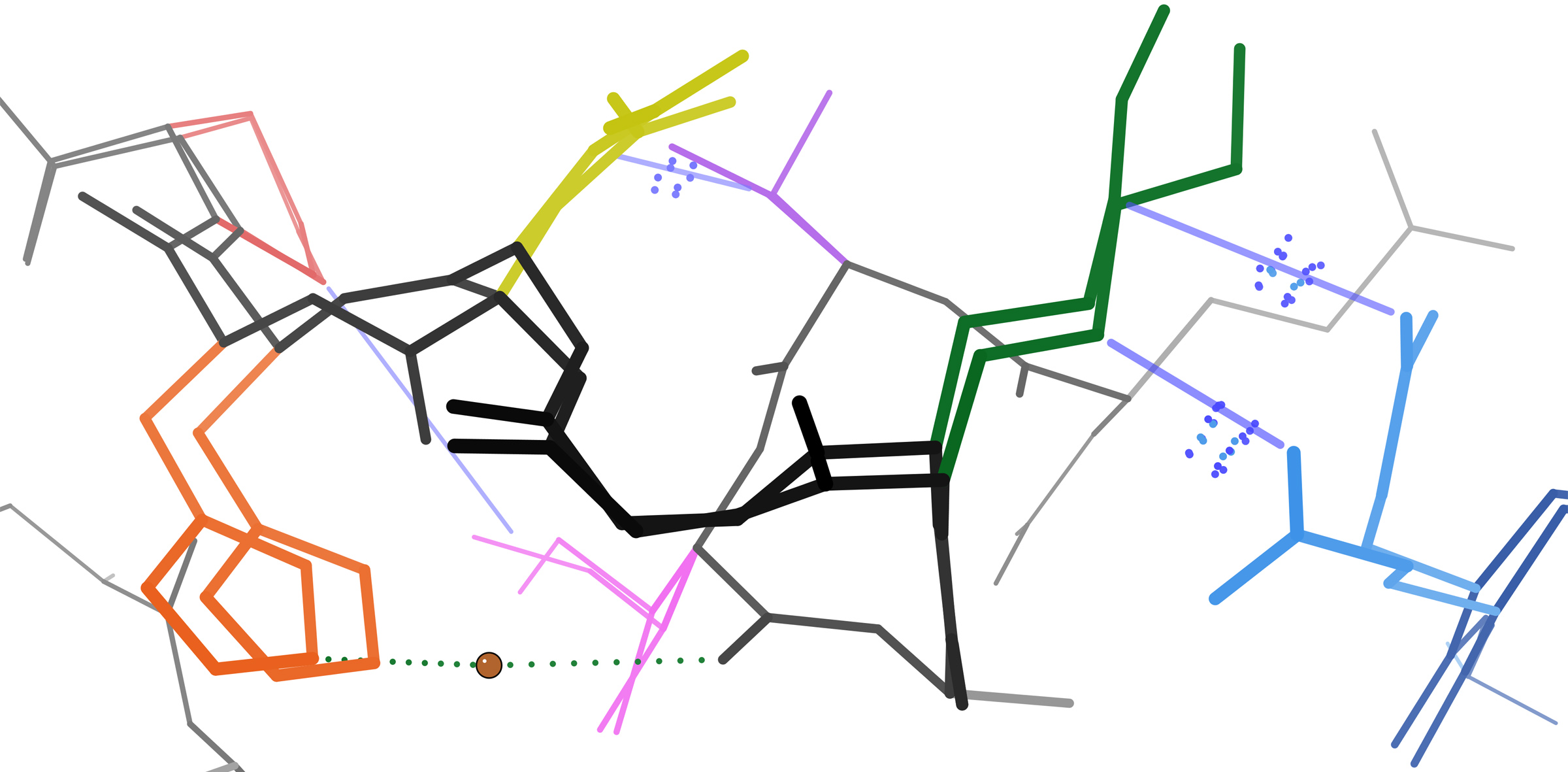
Global Distance Test
The global distance test (GDT), also written as GDT_TS to represent "total score", is a measure of similarity between two protein structures with known amino acid correspondences (e.g. identical amino acid sequences) but different tertiary structures. It is most commonly used to compare the results of protein structure prediction to the experimentally determined structure as measured by X-ray crystallography, protein NMR, or, increasingly, cryoelectron microscopy. The GDT metric was developed by Adam Zemla at Lawrence Livermore National Laboratory and originally implemented in the Local-Global Alignment (LGA) program. It is intended as a more accurate measurement than the common root-mean-square deviation (RMSD) metric - which is sensitive to outlier regions created, for example, by poor modeling of individual loop regions in a structure that is otherwise reasonably accurate. The conventional GDT_TS score is computed over the alpha carbon atoms and is reported as a percentage, r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a ''residue'', which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha And Beta Carbon
In the nomenclature of organic chemistry, a locant is a term to indicate the position of a functional group or substituent within a molecule. Numeric locants The International Union of Pure and Applied Chemistry (IUPAC) recommends the use of numeric prefixes to indicate the position of substituents, generally by identifying the parent hydrocarbon chain and assigning the carbon atoms based on their substituents in order of precedence. For example, there are at least two isomers of the linear form of pentanone, a ketone that contains a chain of exactly five carbon atoms. There is an oxygen atom bonded to one of the middle three carbons (if it were bonded to an end carbon, the molecule would be an aldehyde, not a ketone), but it is not clear where it is located. In this example, the carbon atoms are numbered from one to five, which starts at one end and proceeds sequentially along the chain. Now the position of the oxygen atom can be defined as on carbon atom number two, three o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Template Modeling Score
In bioinformatics, the template modeling score or TM-score is a measure of similarity between two protein structures. The TM-score is intended as a more accurate measure of the global similarity of full-length protein structures than the often used RMSD measure. The TM-score indicates the similarity between two structures by a score between (0,1], where 1 indicates a perfect match between two structures (thus the higher the better). Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold. A quantitative study shows that proteins of TM-score = 0.5 have a posterior probability of 37% in the same CATH topology family and of 13% in the same SCOP fold family. The probabilities increase rapidly when TM-score > 0.5. The TM-score is designed to be independent of protein lengths. The TM-score equation TM-score between two protein structures (e.g., a template structure and a target st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dynamics
In molecular biology, proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Transitions between these states occur on a variety of length scales (tenths of angstroms to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. The study of protein dynamics is most directly concerned with the transitions between these states, but can also involve the nature and equilibrium populations of the states themselves. These two perspectives— kinetics and thermodynamics, respectively—can be conceptually synthesized in an " energy landscape" paradigm: highly populated states and the kinetics of transitions between them can be described by the depths of energy wells and the heights of energy barriers, respectively. Local flexibility: atoms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Angstrom
The angstrom (; ) is a unit of length equal to m; that is, one ten-billionth of a metre, a hundred-millionth of a centimetre, 0.1 nanometre, or 100 picometres. The unit is named after the Swedish physicist Anders Jonas Ångström (1814–1874). It was originally spelled with Swedish letters, as Ångström and later as ångström (). The latter spelling is still listed in some dictionaries, but is now rare in English texts. Some popular US dictionaries list only the spelling ''angstrom''. The unit's symbol is Å, which is a letter of the Swedish alphabet, regardless of how the unit is spelled. However, "A" or "A.U." may be used in less formal contexts or typographically limited media. The angstrom is often used in the natural sciences and technology to express sizes of atoms, molecules, microscopic biological structures, and lengths of chemical bonds, arrangement of atoms in crystals, wavelengths of electromagnetic radiation, and dimensions of integrated circuit part ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Carbon
In the nomenclature of organic chemistry, a locant is a term to indicate the position of a functional group or substituent within a molecule. Numeric locants The International Union of Pure and Applied Chemistry (IUPAC) recommends the use of numeric prefixes to indicate the position of substituents, generally by identifying the parent hydrocarbon chain and assigning the carbon atoms based on their substituents in order of precedence. For example, there are at least two isomers of the linear form of pentanone, a ketone that contains a chain of exactly five carbon atoms. There is an oxygen atom bonded to one of the middle three carbons (if it were bonded to an end carbon, the molecule would be an aldehyde, not a ketone), but it is not clear where it is located. In this example, the carbon atoms are numbered from one to five, which starts at one end and proceeds sequentially along the chain. Now the position of the oxygen atom can be defined as on carbon atom number two, three o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups ( alpha- , beta- , gamma- amino acids, etc.); other categories relate to polarity, ionization, and side-chain group type ( aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino-acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence. Amino acids are formally named by the IUPAC- IUBMB Joint Commi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Side Chain
In organic chemistry and biochemistry, a side chain is a substituent, chemical group that is attached to a core part of the molecule called the "main chain" or backbone chain, backbone. The side chain is a hydrocarbon branching element of a molecule that is attached to a larger hydrocarbon backbone. It is one factor in determining a molecule's properties and reactivity. A side chain is also known as a pendant chain, but a pendant group (side group) has a different definition. Conventions The placeholder R is often used as a generic placeholder for alkyl (saturated hydrocarbon) group side chains in structural formulae. To indicate other non-carbon groups in structure diagrams, X, Y, or Z are often used. History The ''R'' symbol was introduced by 19th-century French chemist Charles Frédéric Gerhardt, who advocated its adoption on the grounds that it would be widely recognizable and intelligible given its correspondence in multiple European languages to the initial letter of "r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CASP
Critical Assessment of Structure Prediction (CASP), sometimes called Critical Assessment of Protein Structure Prediction, is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994. CASP provides research groups with an opportunity to objectively test their structure prediction methods and delivers an independent assessment of the state of the art in protein structure modeling to the research community and software users. Even though the primary goal of CASP is to help advance the methods of identifying protein three-dimensional structure from its amino acid sequence many view the experiment more as a "world championship" in this field of science. More than 100 research groups from all over the world participate in CASP on a regular basis and it is not uncommon for entire groups to suspend their other research for months while they focus on getting their servers ready for the experiment and on performing the detailed predictio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Loop (biochemistry)
A turn is an element of secondary structure in proteins where the polypeptide chain reverses its overall direction. Definition According to one definition, a turn is a structural motif where the Cα atoms of two residues separated by a few (usually 1 to 5) peptide bonds are close (less than ). The proximity of the terminal Cα atoms often correlates with formation of an inter main chain hydrogen bond between the corresponding residues. Such hydrogen bonding is the basis for the original, perhaps better known, turn definition. In many cases, but not all, the hydrogen-bonding and Cα-distance definitions are equivalent. Types of turns Turns are classified according to the separation between the two end residues: * In an α-turn the end residues are separated by ''four'' peptide bonds (''i'' → ''i'' ± 4). * In a β-turn (the most common form), by ''three'' bonds (''i'' → ''i'' ± 3). * In a γ-turn, by ''two'' bonds (''i'' → ''i'' ± 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid Sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the encoded 22, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically synthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Outlier
In statistics, an outlier is a data point that differs significantly from other observations. An outlier may be due to a variability in the measurement, an indication of novel data, or it may be the result of experimental error; the latter are sometimes excluded from the data set. An outlier can be an indication of exciting possibility, but can also cause serious problems in statistical analyses. Outliers can occur by chance in any distribution, but they can indicate novel behaviour or structures in the data-set, measurement error, or that the population has a heavy-tailed distribution. In the case of measurement error, one wishes to discard them or use statistics that are robust statistics, robust to outliers, while in the case of heavy-tailed distributions, they indicate that the distribution has high skewness and that one should be very cautious in using tools or intuitions that assume a normal distribution. A frequent cause of outliers is a mixture of two distributions, wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |